Exploratory applications
At Lunaphore, we are innovators, striving to push the boundaries of research and scientific discoveries.
Lunaphore’s research team regularly works on exploratory projects and applications, with the aim of developing tools that can address a variety of new research questions.
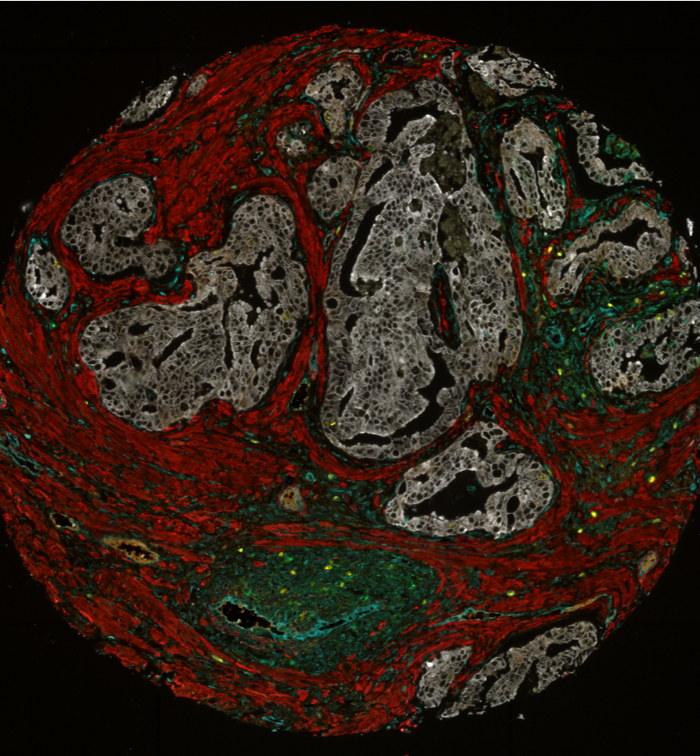
Signal amplification to boost marker detection
The sequential immunofluorescence (seqIF™) technique on COMET™ currently offers linear amplification with the use of secondary antibodies, enabling the efficient detection of multiple markers, preserving the quantification possibilities of antigen expression and its heterogeneity, and avoiding saturation.
New investigative data showed that additional signal-boosting techniques can be optionally used on COMET™ and integrated into hyperplex seqIF™ assays.
These findings will be useful when a strong, non-linear, and only qualitative detection of makers is preferred.
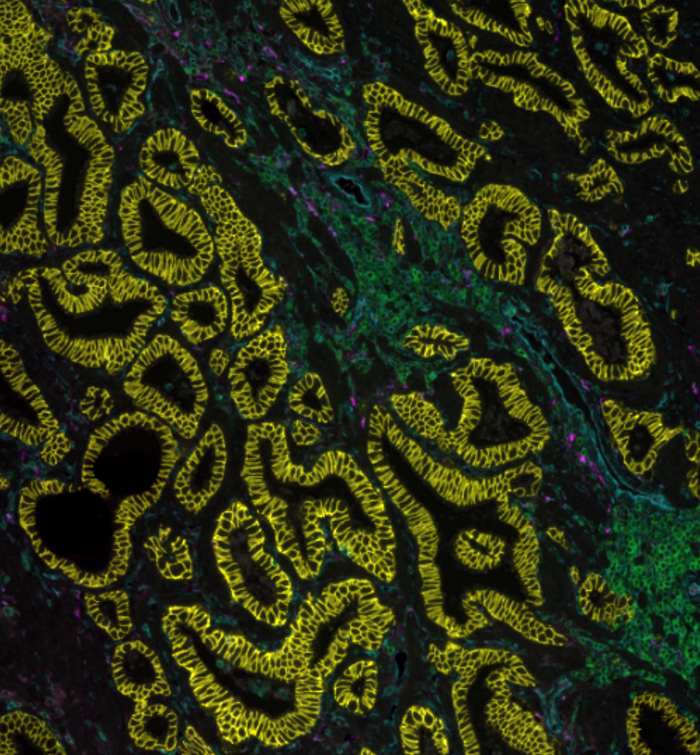
Why signal boosting:
1. To detect markers when a qualitative analysis is preferred over a quantitative one.
2. To amplify the detection of low-expressed or difficult targets.
3. To expand the number of detectable targets when testing new or non-validated markers.
4. To reduce antibody consumption and to increase throughput