Spatial Biology Education
Advancing efficient antibody elution, epitope integrity and tissue preservation on the COMET™ platform
Posted on:
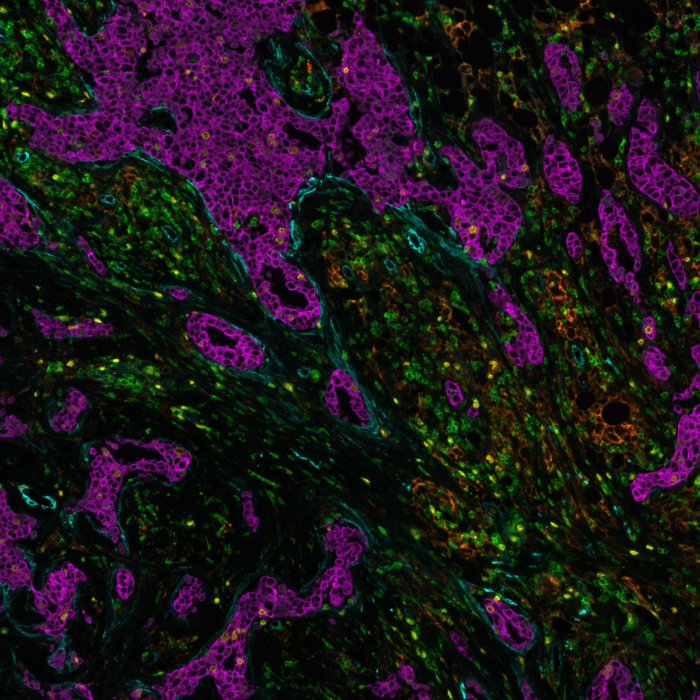
Spatial proteomics is a rapidly evolving field that allows the detection of proteins within biological tissues. The simultaneous detection of protein localization and expression helps scientists and clinicians gain deeper insights into the molecular and cellular interactions within complex tissues. Multiplex immunofluorescence (mIF) is a well-established method in spatial proteomics that enables the detection of multiple protein biomarkers simultaneously on the same tissue section.
Designing a robust and reliable multiplex proteomics experiment requires careful planning to ensure successful detection of multiple protein biomarkers, while maintaining sample integrity. Lunaphore’s Fast Fluidic Exchange (FFeX™) technology is a cutting-edge microfluidics platform that streamlines antibody-based staining, imaging and elution, allowing the efficient detection of protein targets while preserving tissue and epitope stability. The FFeX™ technology enables a rapid immunostaining workflow by drastically reducing incubation times, while maintaining high antigen detection efficiency, resulting in robust and consistent spatial biomarker detection.
The seqIF™ workflow
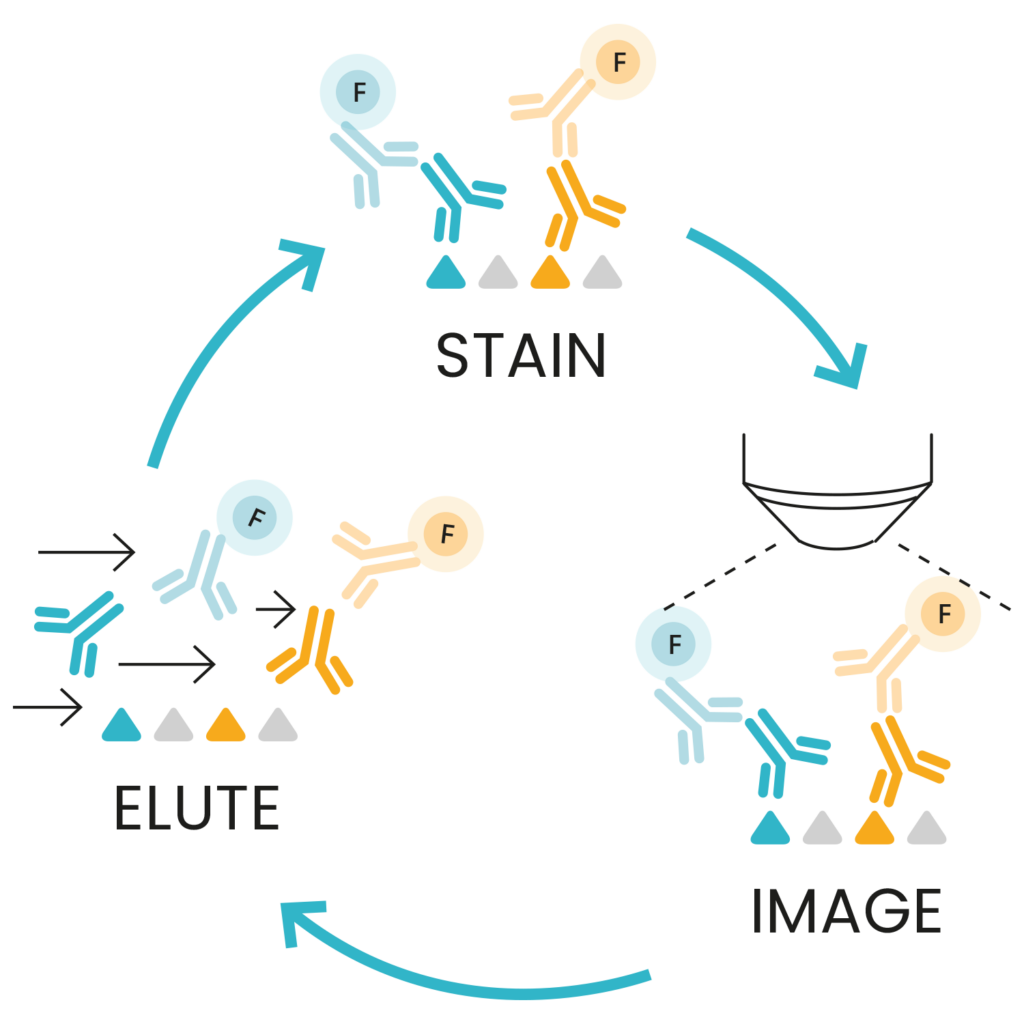
Sequential immunofluorescence1 (seqIF™) is an innovative technology with the potential to transform hyperplex spatial proteomics. seqIF™ provides a streamlined alternative to lengthy workflows that require labor-intensive and costly antibody customization through barcoding or conjugation. seqIF™ also addresses reproducibility challenges and tissue damage due to repetitive staining cycles associated with cyclic immunofluorescence methods.
The seqIF™ workflow is based on repeated sequences of staining, imaging, and antibody elution on the same tissue section as shown in Figure 1. This process utilizes Lunaphore’s FFeX™ technology in use with proprietary reagents and it is designed to maximize the number of detectable targets, while preserving tissue integrity, tissue morphology, and epitope stability, ensuring reliable and consistent results.
Related Articles
A high-plex road to biomarker development in a fully automated workflow
Posted on 22 Jun 2023
Read PostThe role of spatial biology in studying inflammation: A novel dimension
Posted on 02 Aug 2023
Read Post