Webinar
Immuno-oncology: look beneath the surface with multiplex sequential immunofluorescence (seqIF™)
Posted on:
The rise of spatial biology enhances the study of the tumor microenvironment (TME) in immuno-oncology
What proteins? Where? When? – are the major questions that every scientist asks to generate a comprehensive picture of protein expression. The study of the spatial distribution of cells and their microenvironment is known as spatial biology, growing field of interest in immuno-oncology research. The interactions between various cells within the tumor microenvironment (TME) are important to investigate from a molecular standpoint. Spatial biology allows researchers to examine the distance between cells, decipher expression of molecules and generate cell density metrics. As highlighted by Dr. Paolo Ascierto, Director of the Department of Melanoma, Cancer Immunotherapy and Development Therapeutics at the National Tumor Institute in Naples, “for immuno-oncologists such knowledge is crucial to understanding the process that underlies the immune response to treatment”1.
Intensive research is currently being conducted by scientific community to identify predictors of response to anti-PD-1/PD-L1 (anti–programmed cell death 1/programmed cell death ligand 1) immunotherapy helping to match patients to the right treatment. Such discoveries also drive the target identification and development of new drugs.
What are the common challenges faced when starting off in spatial biology?
In recent years, techniques used in spatial biology have advanced substantially. Particularly spatial biology proteomics with visualization of multiple markers on the same tissue has raised considerable interest. However, for those just starting in this field, this can present several hurdles, including:
- Extensive hands-on time due to lengthy and complex protocol requirements that need substantial optimization and numerous manual steps.
- Tissue morphology degradation due to long exposure times to harsh protocol conditions.
- Lack of flexibility due to the limited offer of proprietary antibody chemistries.
- Technical limitations on the number of antigens that can be visualized at the same time depending on the chosen chemistry or the number of microscope channels.
- Limited sample availability.
Which techniques should you try first to ease adoption?
All the above-mentioned hurdles can seem daunting at first but there are some methods that are easier than others.
Researchers have a wide range of tools available to study spatial biology. Some of the existing approaches for multiple biomarker detection are the following:
- Tyramide Signal Amplification (TSA), where reagents are incubated sequentially but visualized simultaneously.
- Antibody mixes, for lower-plex stainings, where antibodies are incubated together for simultaneous imaging.
- Antibody conjugates, which involve tagging antibodies for independent detection with oligonucleotides, metal tags or breakable fluorophores.
- Sequential immunofluorescence (seqIF™) which relies on standard antibodies that are incubated and eluted in a sequential manner.
Thus, scientists need to evaluate several factors to choose the best technology that supports their experimental objectives.
Sequential immunofluorescence (seqIF™) is a novel and simple approach for the detection of multiple proteins on a single slide with minimal optimization efforts. The first step consists of staining a tissue section by incubating it with primary antibodies, followed by a marker detection step with fluorescently labelled secondary antibodies. Then, the images are acquired through an imaging step, followed by an elution step, which is the removal of the antibodies with elution buffer (Figure 1). Subsequently, another round of staining can be conducted to visualize a new set of antigens. Antibodies from different species can be used within each cycle. Then, the acquired images can be stacked to create overlays to build spatial relationships maps.
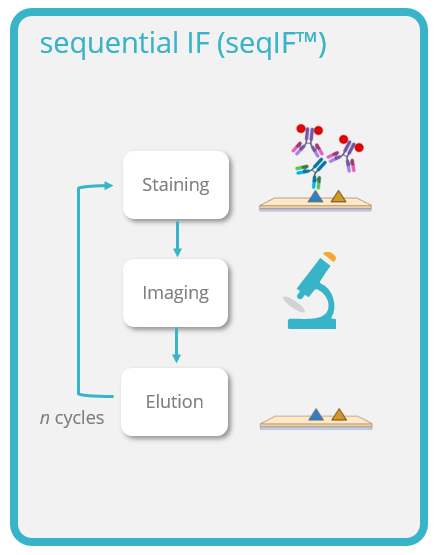
What are the main advantages of seqIF™?
- Time savings: seqIF™ allows you to perform quick multiplex marker optimization and assay development.
- Enhanced flexibility: seqIF™ can be performed on FFPE (formalin-fixed tissues embedded in paraffin) as well as on FS (frozen sections) with customized protocols. Antibodies from the same host can be used at distinct staining steps.
- Increased multidimensional insights: obtain the most comprehensive data out of your precious samples allowing you to visualize multiple antigens at the same time that illuminate cellular microenvironments.
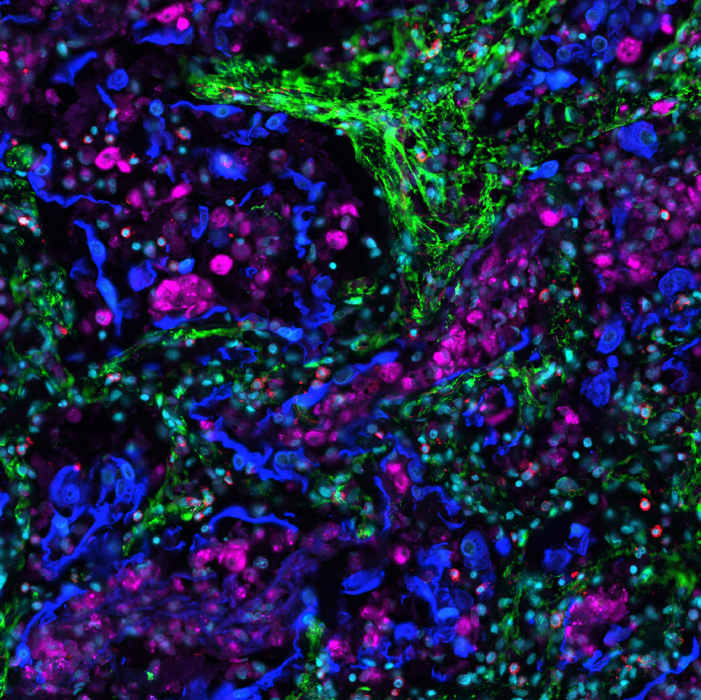
As the method is easily scalable, seqIF™ can be particularly useful in the design of immunophenotyping panels where testing of multiple antibodies is required. It allows you to visualize protein co-expression on immune cell subsets to investigate their spatial correlations with functional significance (Figure 2).
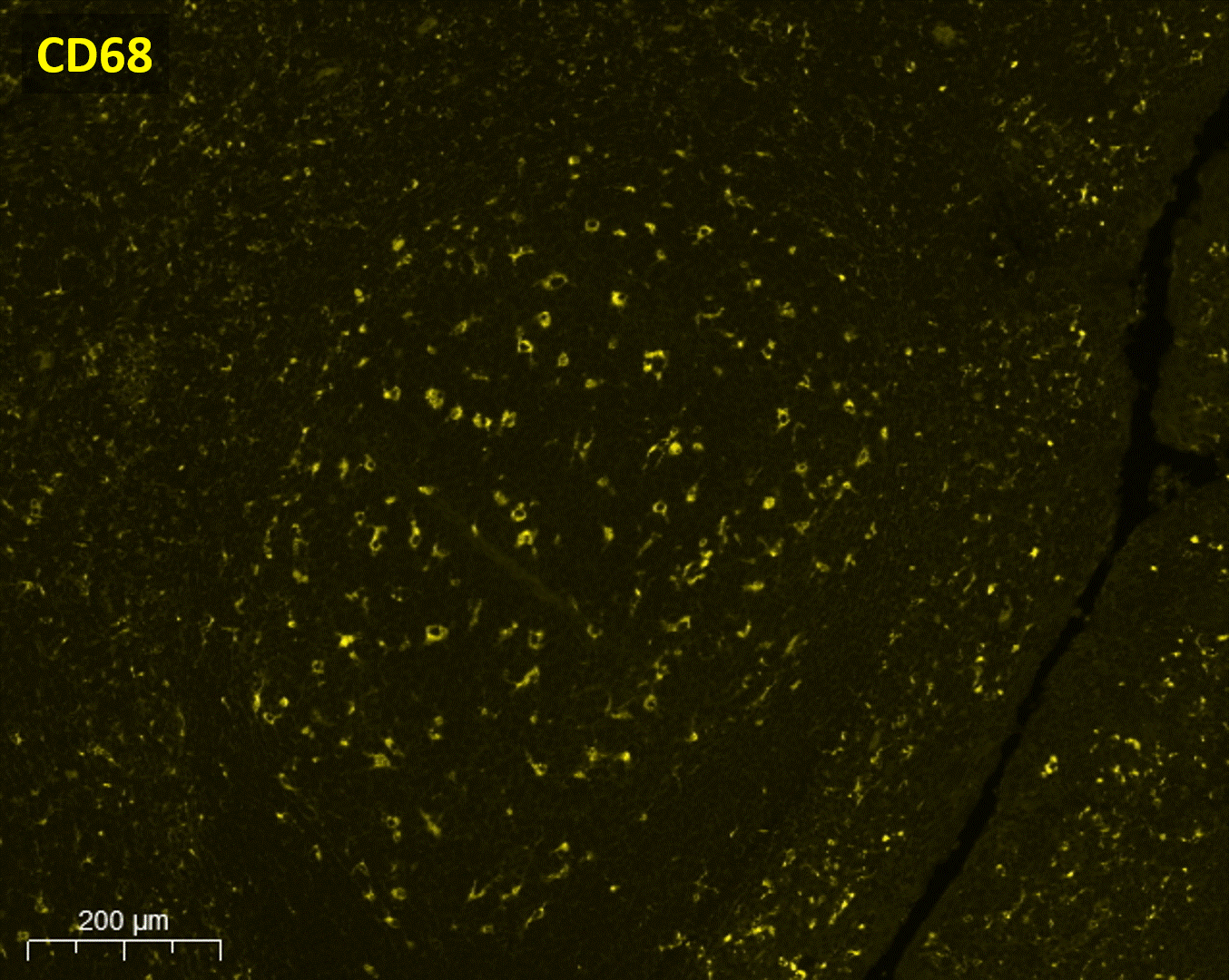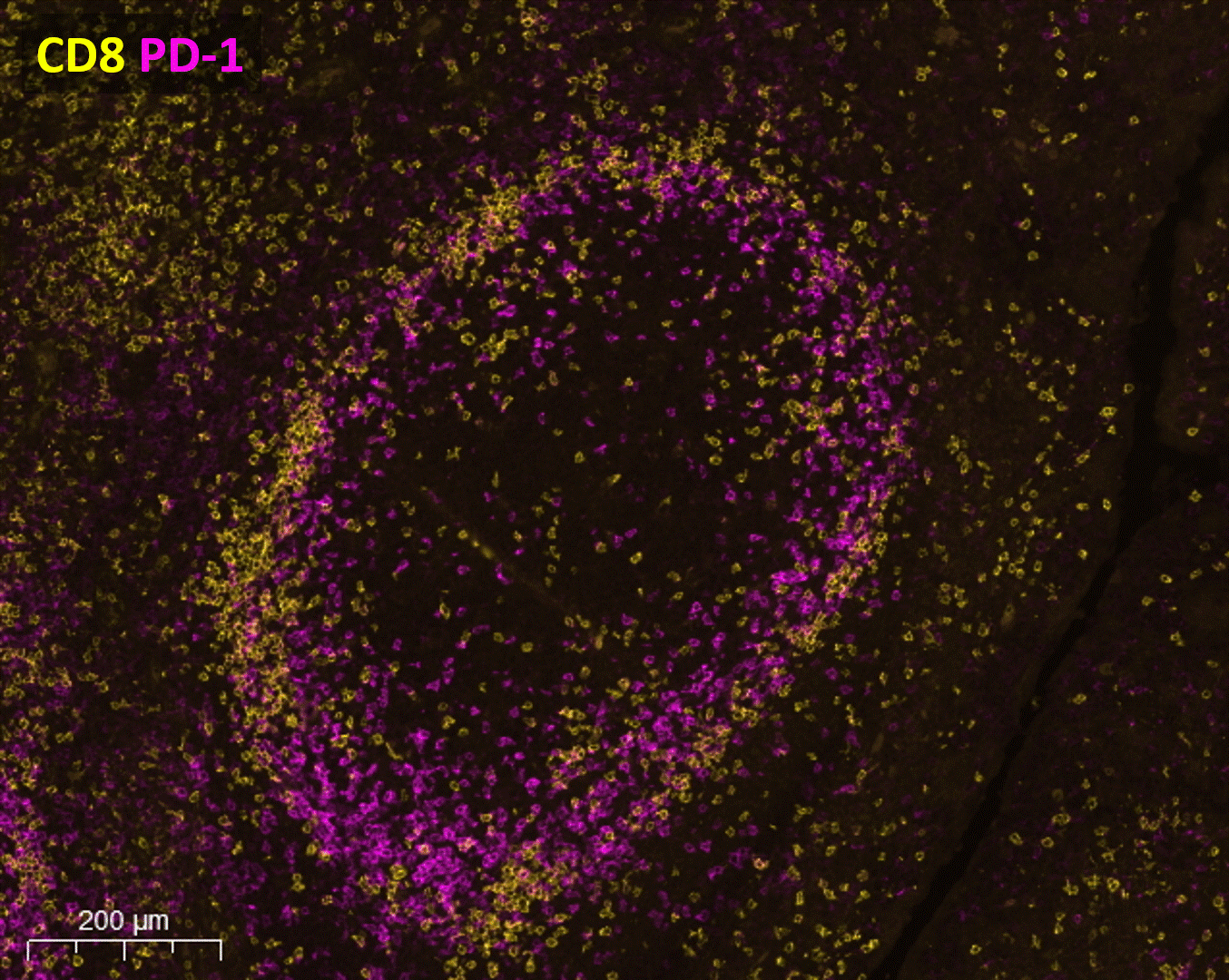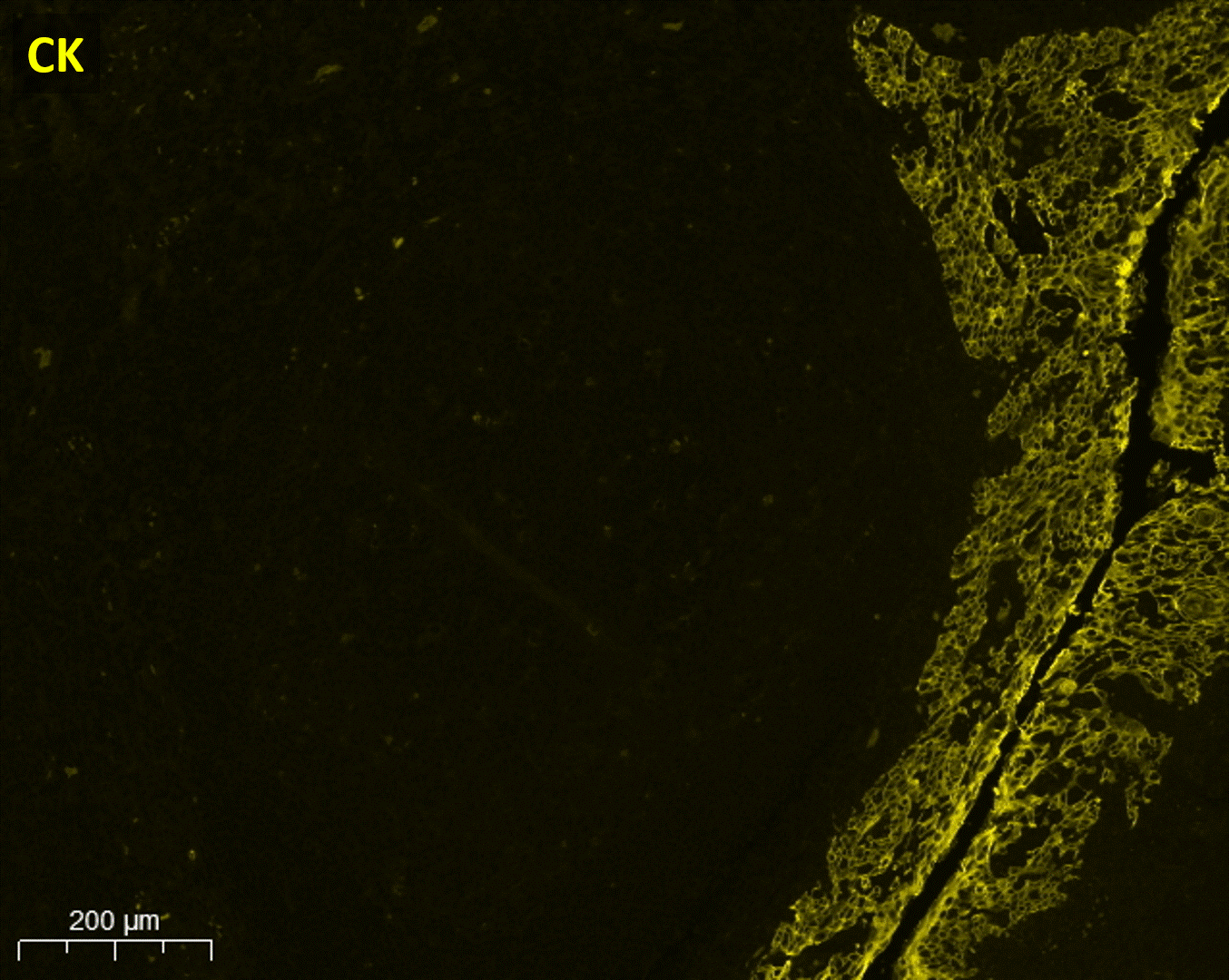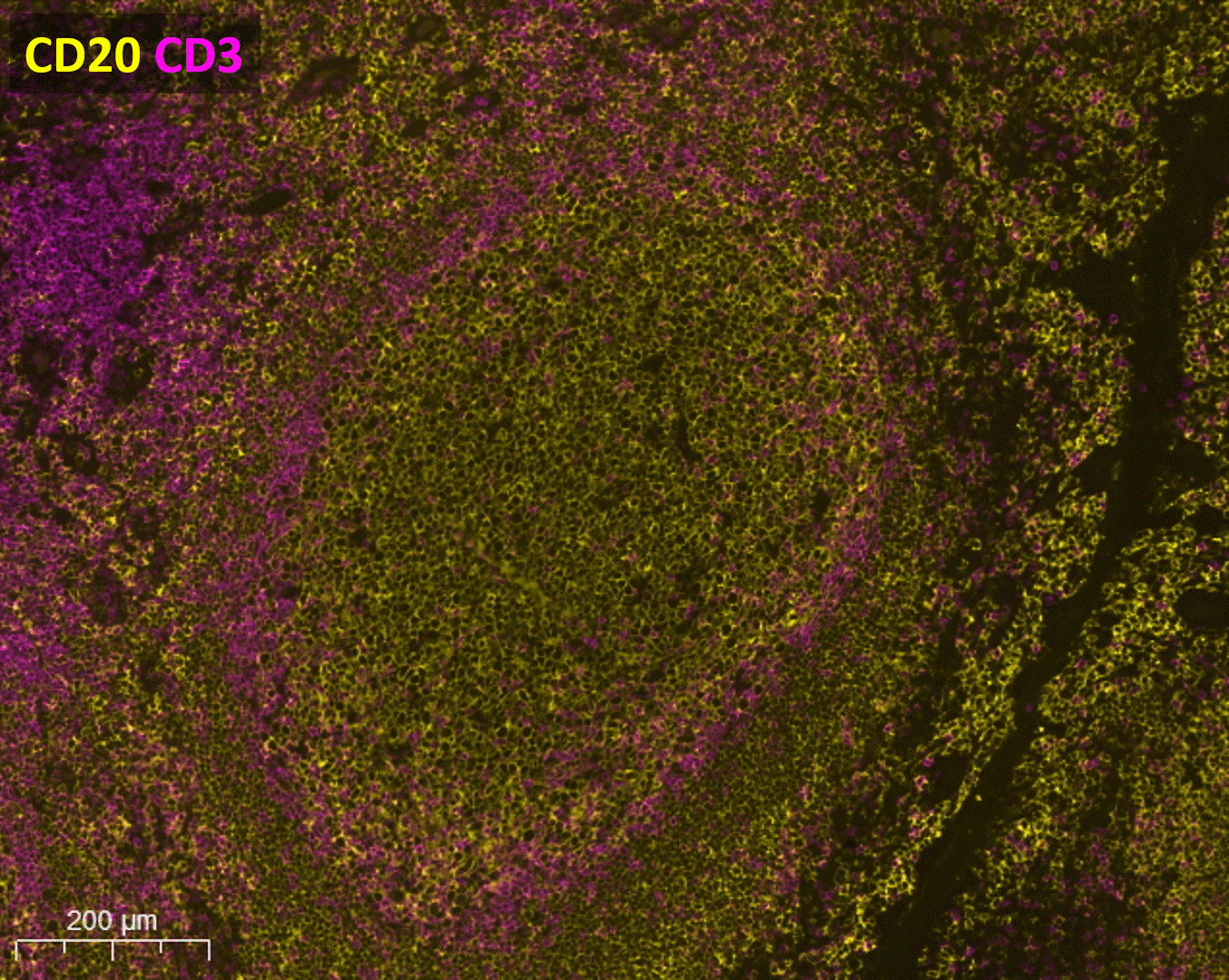
Available automated solution
Several automated tools for looking into the spatial context are available and in order to choose the finest technology for a lab, each researcher must examine a number of aspects. Lunaphore Technologies offers a unique solution, designed to address spatial biology hurdles with one instrument, LabSat®. The core instrument technology called Fast Fluidic Exchange (FFeX), is based on a microfluidic “Staining Chip” sequentially delivering reagents onto a tissue sample in a controlled manner.
Integrating LabSat® in your lab allows for:
- Fast generation of high-quality data: thanks to Lunaphore’s microfluidic technology and a pressurized reagent delivery system, incubation times are greatly reduced.
- High tissue preservation: LabSat®’s short incubation times limit the exposure of the tissue to harsh conditions, which allows to preserve tissue morphology and epitope stability, providing you with consistent and reliable data despite multiple rounds of elution.
- Spatial biology with your standard, non-conjugated antibodies: perform your experiments with your antibodies of choice, and leverage the libraries you have previously characterized.
- Quick and easy optimization: create your customized protocols in a blink of an eye due to the easy and trouble-free user interface.
On one single platform LabSat® researchers can ask sophisticated questions to extract a multitude of information from valuable samples allowing them to see what’s happening in the tissue microenvironment. If you are a small or medium lab looking into automating spatial biology workflows LabSat® is the right solution for your needs.
References:
1 Spatial Biology: Transforming Immuno-Oncology Research. The Pathologist. Available from: https://thepathologist.com/diagnostics/spatial-biology-transforming-immuno-oncology-research [Accessed 11.02.2022].