Spatial Biology Education
Uncovering the mechanisms behind immune evasion in pancreatic cancer with multiplex immunofluorescence
Posted on:
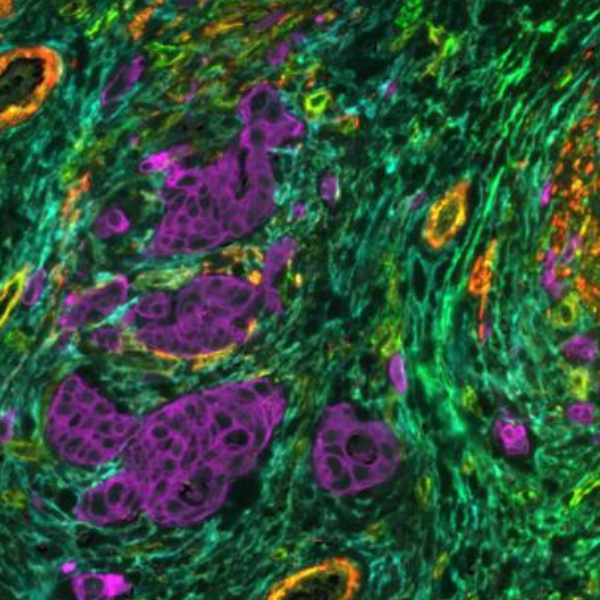
Pancreatic ductal adenocarcinoma (PDAC) is a highly aggressive, fatal cancer1. The typical 5-year survival rate for PDAC tumors is less than 10%, despite scientific advancements in understanding the disease’s biology and the creation of novel therapeutic approaches2. Immune cells play a crucial role in detecting and destroying abnormal cells, but cancer cells can sometimes evade or suppress the immune response. An improved understanding of the tumor immune microenvironment (TME) within PDAC tumors is needed to establish effective immunotherapy treatments. It is known for its highly desmoplastic stroma, an increased growth of connective tissue (or stroma) that happens as a response of the host tissue to invasive cancer cells. Desmoplastic stroma plays a critical role in supporting tumor growth and progression and is thought to contribute to the resistance of PDAC to various therapies. Spatial biology has shown great promise in providing biological insights about this process.
Multiplex immunofluorescence (mIF) as an essential tool in the understanding of PDAC
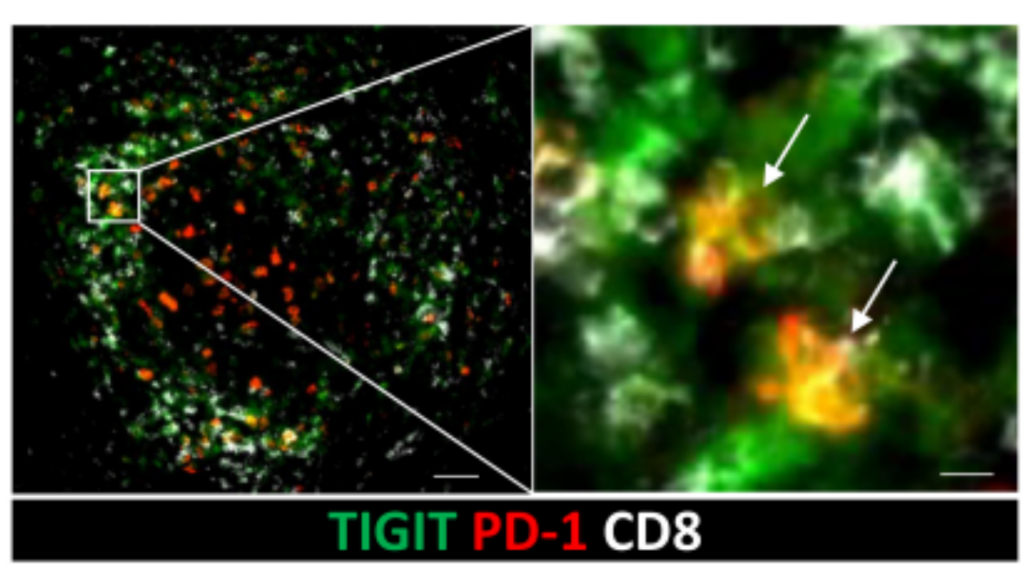
Immune checkpoints are proteins found on the surface of immune cells, such as T cells, and interact with other proteins called ligands, which are found on the surface of target cells, such as some tumor cells. When an immune checkpoint is blocked, it can enhance the immune response, allowing the immune system to attack and eliminate cancer cells. A recent study used mIF, cytometry by time of flight, and single-cell RNA sequencing to characterize tumor-infiltrating T-cell populations in PDAC patients3. Large populations of CD8 expressing tissue-resident memory T cells (TRM) were found. TRMs are a type of T cell that provides a rapid and localized response to antigen challenge4. Antigen challenge refers to the process of exposing the immune system to a specific antigen, which is a molecule that can be recognized by the immune system, such as a protein. These TMRs were expressing PD-1 and TIGIT, two immune checkpoint proteins, at high levels. This discovery was facilitated by performing multiplex immunofluorescence on COMET™ to determine the precise co-localization of two checkpoint proteins from the same TRM cells (Figure 1).
Further authors examined the expression of the ligands for PD-1 (PD-L1 and PD-L2) and TIGIT (CD155 and CD112) on cells within the TME. In ectopic lymphoid structures, PD-L1, PD-L2, and CD155 were expressed in the T-cell region and colocalized with PD-1+TIGIT+CD8+T cells3 (Figure 2).
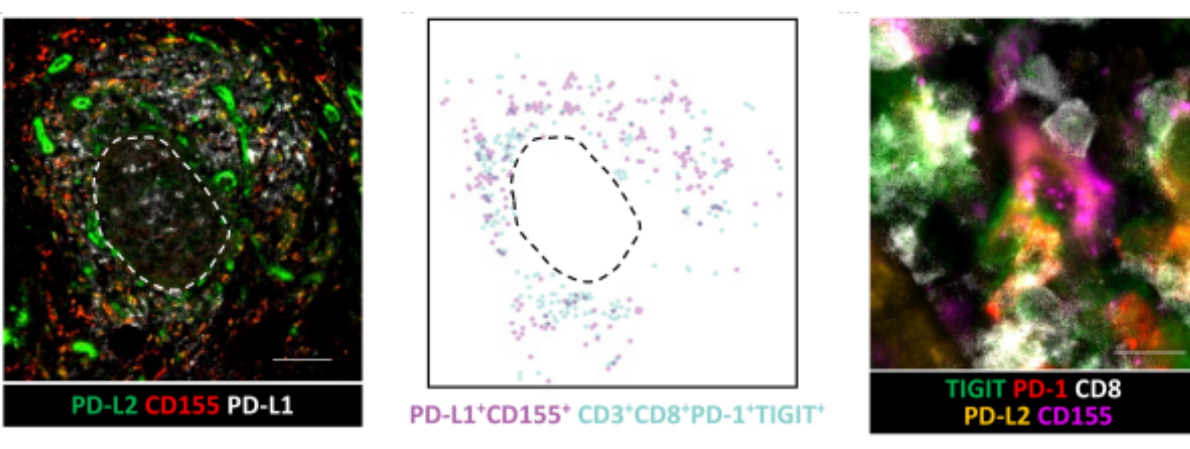
PD-1 and TIGIT suppress the activity of T cells, which may contribute to the cancer cells’ ability to evade the immune response. However, the study found that blocking both PD-1 and TIGIT with antibodies can reverse this inhibition and restore the activity of TRM cells. Overall, this research result demonstrates a potential strategy for enhancing the immune response against PDAC consisting in targeting the PD-1 and TIGIT checkpoint proteins on TRM cells3.
Related Articles
The Spatial Biology Week™ 2022: the transforming power of spatial biology
Posted on 07 Nov 2022
Read Post