Spatial Biology Education
A high-plex road to biomarker development in a fully automated workflow
Posted on:
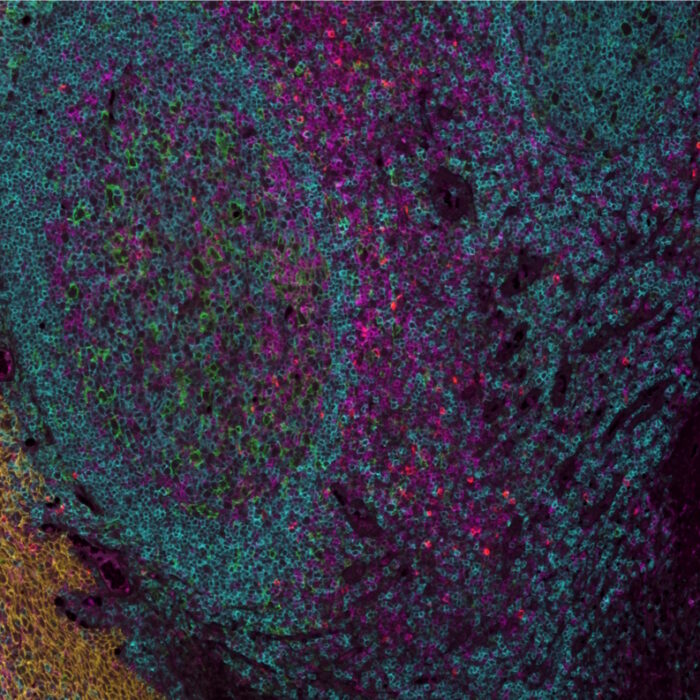
Many biological processes are driven by the interactions and communication between different cell types, as well as the spatial arrangement of these cells within tissues and organs. Spatial biology explores these relationships and neighborhoods with the aim to uncover complex biological processes and provide mechanistic hypotheses on disease development and therapy resistance. Especially high-plex, or high-plexity in spatial biology allows for the simultaneous detection and measurement of multiple molecular targets within the same sample. Such techniques have become highly valuable in the biomarker development field for example, where bulk analyses have reached a bottle neck. Therefore, researchers are pursuing novel biomarker signatures that capture the spatial context using multiplexed assays. Feng et al.1 highlight the potential of mIHC/IF for multiparametric immune profiling, which may benefit clinical trial stratification of patients with oral squamous cell carcinoma (OSCC). The authors demonstrated that high CD8+ T cell counts near the invasive margin significantly correlate with a better survival rate. Notably, the location of suppressive factors, in addition to their composition, is decisive. Cells that are PD-L1+ or FOXP3+ and close to CD8+ T cells have lower overall survival rates. Such results provide new opportunities to discover novel biomarkers for patients stratification.
Different approaches exist in the field of spatial biology to create these cell-by-cell maps. Some researchers start with a smaller panel of 4-5 antibodies to validate their staining protocol and ensure the robustness of their analysis pipeline before expanding the panel to additional targets. Others may require larger panels of 20 or more antibodies to comprehensively analyze complex biological systems. Creating a multiplex immunofluorescence panel can be a challenging task, especially when handling samples with limited access.
Discerning protein expression using sequential immunofluorescence (seqIF™)
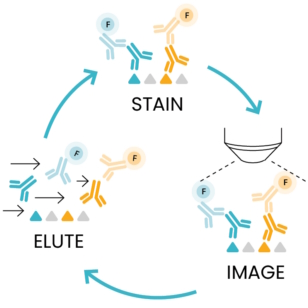
As the number of protein biomarkers identified via multiplexing rises, efforts have been placed to create automated platforms that ensure a high degree of reproducibility and shorten the time required for tissue staining and processing. COMET™, an automated high-throughput instrument developed by Lunaphore, meets such requirements. COMET™ spatial solution uses a novel and simple approach called sequential immunofluorescence. SeqIF™ (Figure 1) consists of sequential cycles of staining, imaging, and elution of 2 markers at a time. This process is repeated 20 times for a total of 40 markers per single run.
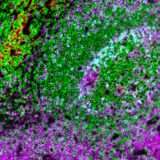
The plex level capability of the technology is virtually unlimited, as it is possible to perform multiple additional hyperplex runs on the same slide using non-conjugated primary antibodies. SeqIF™ approach, which uses controlled, gentle, and rapid incubations, guarantees a well-preserved tissue morphology to obtain hyperplex stainings with high quality and reproducibility. From antigen retrieval to elution, tissues are exposed to mild conditions and only for short incubation times. Figure 2 demonstrates that even after 20 cycles of staining, imaging, and elution, the tissue integrity and epitope stability are preserved, and more than 40 different markers can be accurately detected on the same tissue slide.
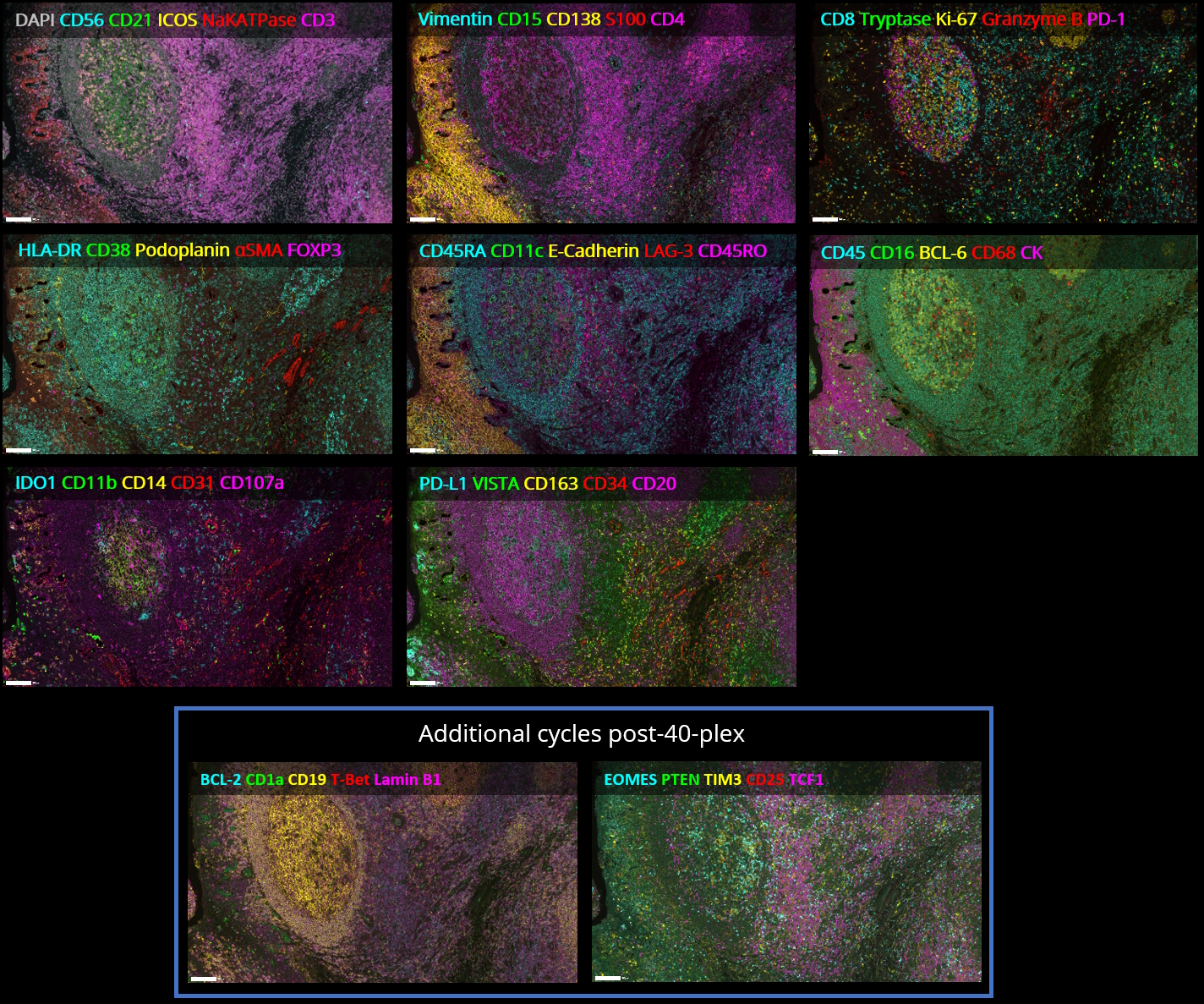
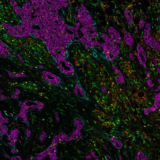
In addition to preserving the tissue integrity, the gentle elution buffer enables an efficient removal of antibodies, without requiring photo- or chemical bleaching. Figure 3 demonstrates that after 40 cycles of seqIF™, tissue integrity and epitope stability are kept. Finally, the same slide can be used to perform any additional downstream modalities, such as H&E (find out more here) or transcriptomics, without the need of tissue transfer steps or additional serial tissue sections.

Obtain large amounts of contextual information
Plex level is becoming increasingly important because it allows researchers to study complex biological systems in a more comprehensive manner. Thanks to novel sequential immunofluorescence approach, it is possible to perform automated, hyperplex analysis with highly preserved tissue morphology allowing for further downstream analysis. The seqIF™ technique has the potential to support and fast track development studies, overcoming technical limitations in spatial biomarker analysis on tissues.
Reference:
- Feng Z et al. Multiparametric immune profiling in HPV– oral squamous cell cancer. JCI Insight. 2017; 2(14):e93652.doi: 10.1172/jci.insight.93652.