Poster
Hyperplex analysis on delicate frozen samples: now possible with high-quality results
Posted on:
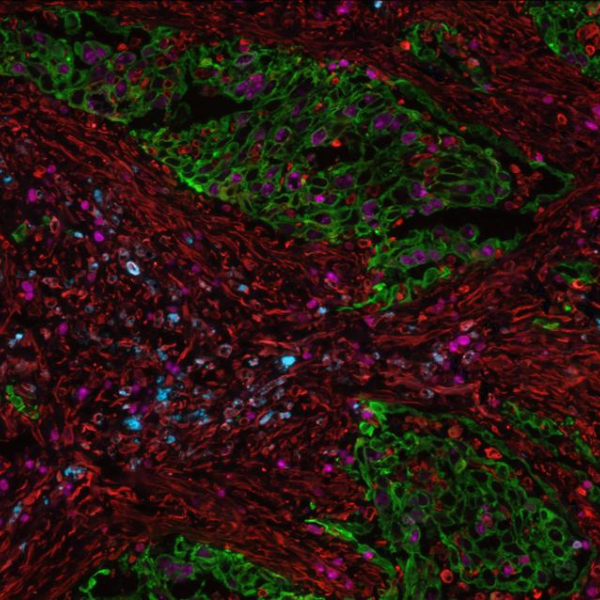
The tumor microenvironment (TME) components significantly impact cancer’s development, growth, metastasis, and the degree to which it responds or resists immunotherapies1, 2. Researchers have been widely using multiplex immunofluorescence (mIF) in recent years because it allows for a multi-layered depiction of the TME. There is a growing need for versatile, high-throughput hyperplex analysis platforms, as translational research studies increase the demand for spatial biomarker analyses3, notwithstanding the possible limited availability and the fragility of the tissue samples. In particular, there is a growing interest from researchers that are profiling TME in analyzing simultaneously multiple biomarkers on frozen sections. The existing manual, error-prone, and time-consuming procedures constrain the usage of this application. Additionally, manual protocols as well as some automated platforms often employ methods that can harm tissue morphology, such as long reagent incubations, high-temperature cycles, or photobleaching steps, which can compromise results.
Discerning protein expression in fragile frozen tissue sections using sequential immunofluorescence (seqIF™)
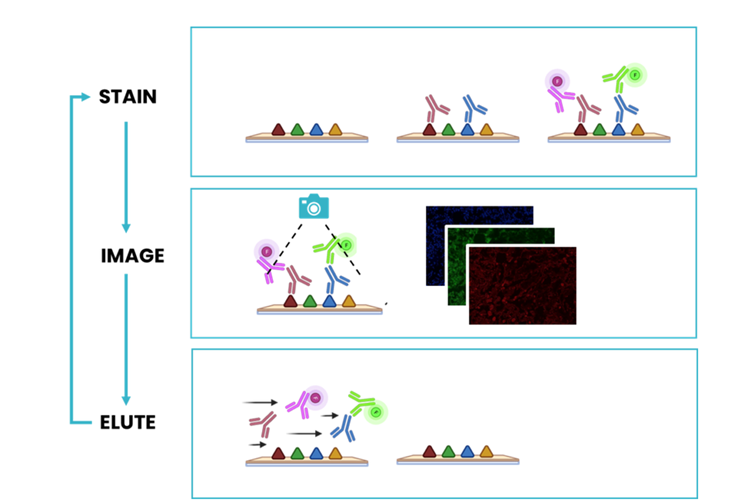
Frozen sections have the benefit of preserving the protein structure, including native antigens in each tissue. They are substantially more delicate than FFPE samples and cannot withstand stringent antigen retrieval procedures. In addition, some tissue types are particularly delicate and require the use of gentle tissue staining protocols in order to preserve tissue morphology. The revolutionary microfluidic-assisted seqIF™ method is compatible with a wide range of tissue types and allows for a comprehensive analysis of delicate samples. It consists of subsequent cycles of mIF staining, imaging, and elution, where two biomarkers are detected simultaneously in every cycle (Figure 1). The COMET™ platform performs fully automated seqIF™ assays with the detection of up to 40 biomarkers, in 20 cycles, on a single tissue slide without user intervention. Thanks to the patented microfluidic chip technology, all conditions are precisely controlled, enabling seqIF™ experiments on COMET™ with exceptional reproducibility, signal uniformity, and highly preserved tissue morphology.
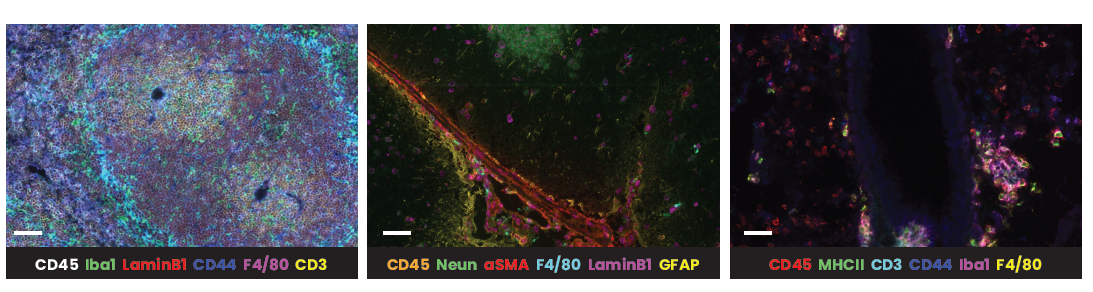
In the example below, murine spleen, brain, and lung tissues were analyzed with multiplex panels demonstrating accurate detection of the specified biomarkers to assess the compatibility of delicate murine samples with the seqIF™ protocol on COMET™ (Figure 2).
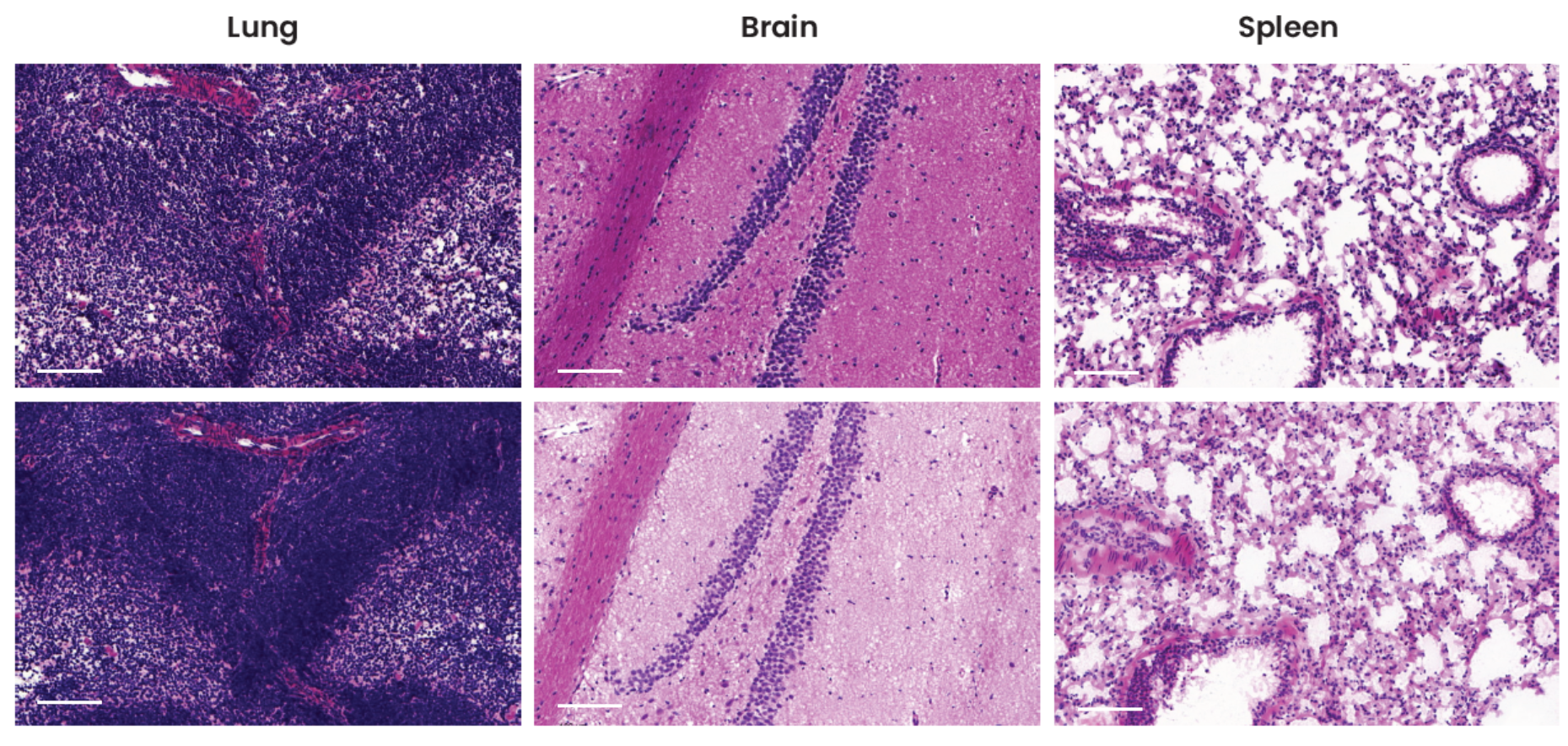
Pathologists have used hematoxylin and eosin (H&E) staining to visualize multiple tissue components in microscopic examinations of biopsies and surgical samples for more than a hundred years4. In addition to mIF, researchers can benefit from using this traditional method on the same slide in order to analyze the tissue architecture and morphological structure. The preservation of tissue morphology is key to performing any additional downstream applications after a multiplex assay, such as H&E or even chromogenic IHC. A study was performed to assess tissue morphology preservation after a seqIF™ protocol. The specimens were further stained with H&E after undergoing a 20-cycle protocol on COMET™, equivalent to a 40-plex analysis. As demonstrated by H&E staining run after seqIF™, as compared to freshly processed control slides, the tissue morphology for all three tissue types was well preserved (Figure 3).
Hyperplex seqIF™ on human lung cancer frozen sections
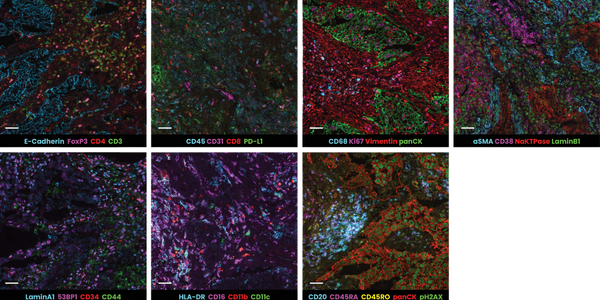
Robust tools are required to better understand the mechanisms underpinning human disease. Some organs present a delicate tissue morphology necessary to achieve their physiological function. When studying the cellular composition of tissue sections from such organs, it is particularly challenging to preserve their native morphology and achieve an accurate representation of the living organ. Using seqIF™ for deep characterization of human lung cancer, 32 biomarkers, including tumor, stroma, and immune cell markers, were simultaneously detected on a single frozen section slide of non-small cell lung cancer (NSCLC) with a total processing time of 23 hours.
COMET™ platform and guided workflow design allowed to optimize protocol in a few steps, which resulted in robust detection of all targets independently of their subcellular localization (Figure 4).
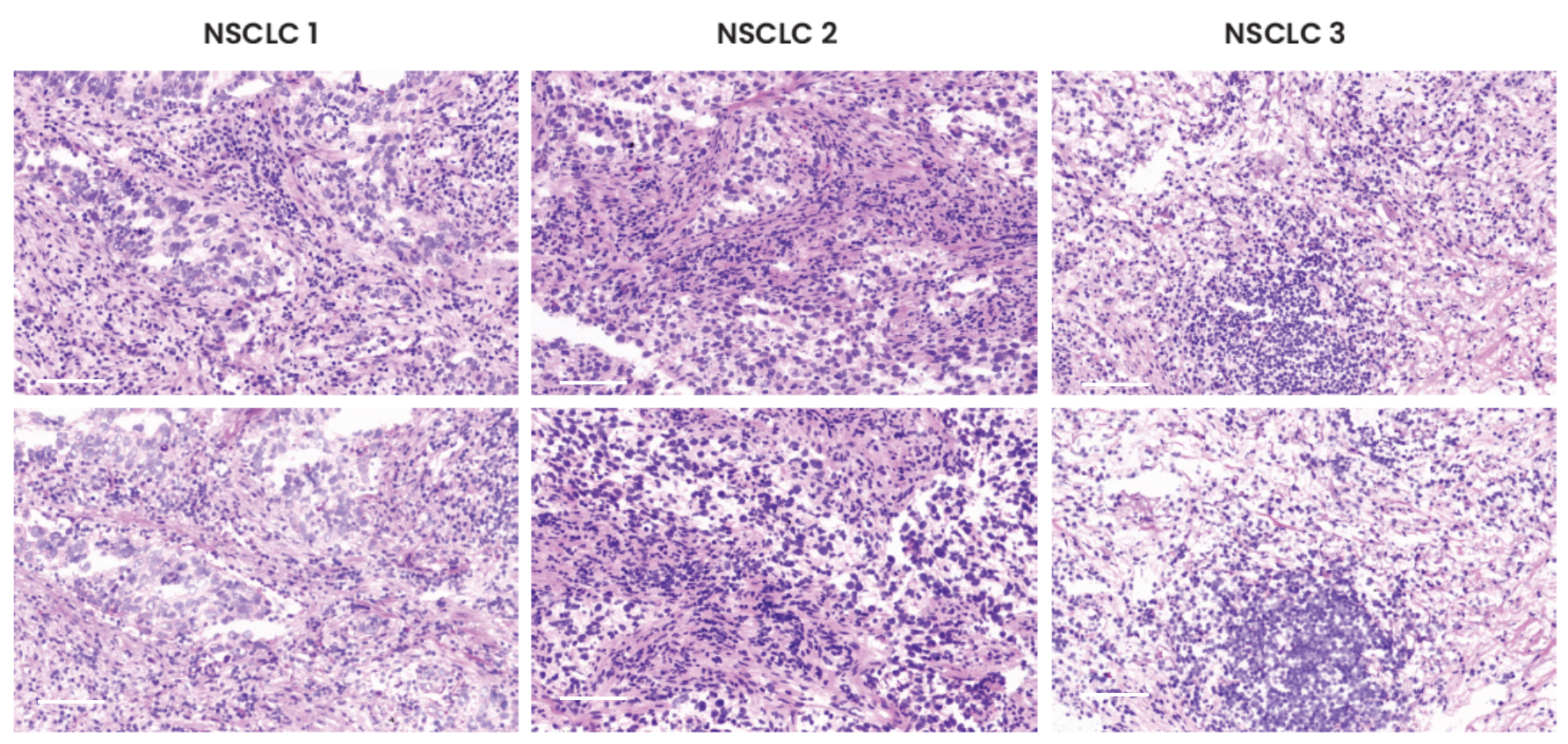
H&E staining was performed on the same slide retrieved from the platform, after running the 16-cycle hyperplex protocol. The results showed well-preserved tissue architecture compared to the unprocessed control tissue (Figure 5).
The elimination of limitations in spatial tissue analysis with seqIF™
Implementation of seqIF™ with the COMET™ platform is compatible with a wide range of tissue types, including delicate tissues, with excellent preservation of tissue morphology. Furthermore, seqIF™ enables downstream analysis with H&E, simplifying the integration of fluorescence imaging with traditional histopathological analysis. The seqIF™ technique has the potential to support and expedite discovery studies across multiple research fields, overcoming technical limitations in spatial biomarker analysis on tissues.
References:
- Hanahan D. and Weinberg R. Hallmarks of cancer: the next generation. Cell. 2011; 144(5):646-74. doi: 10.1016/j.cell.2011.02.013.
- Tumeh P. et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature. 2014; 515(7528):568-71. doi: 10.1038/nature13954.
- Sadeghi Rad H. et al. The evolving landscape of predictive biomarkers in immuno‐oncology with a focus on spatial technologies. Clin Transl Immunology 2020; 9(11):e1215. doi: 10.1002/cti2.1215. eCollection.
- Gurcan MN, Boucheron LE, Can A, Madabhushi A, Rajpoot NM, Yener B. Histopathological Image Analysis: A Review. IEEE Rev BioMed Eng. 2009; 2:147–71. doi: 10.1109/RBME.2009.2034865