Technical Note
Oncology: expanding the research frontier with revolutionary spatial proteomics
Posted on:
The evolution of cancer research
Cancer research has evolved remarkably over the past decades. Since the 2000s various techniques have been used to study tumors including bulk genomic and single-cell analyses (e.g., RNA sequencing)1. These approaches have been critical in the discovery of new biomarkers, the molecular regulation of tumor growth, and the molecular drivers of metastasis and drug resistance. However, their limitations lie in their inability to provide a full image of the tumor microenvironment (TME) and cellular location of these biomarkers2. It is therefore important to not only investigate the broad changes in tumor biology but also to gain insights into the molecular variation of cells (heterogeneity within [intra-] and between [inter-] tumors), to improve the diagnosis and treatment of specific cancer subtypes and to develop more personalized therapies1.
The emergence of immuno-oncology therapies
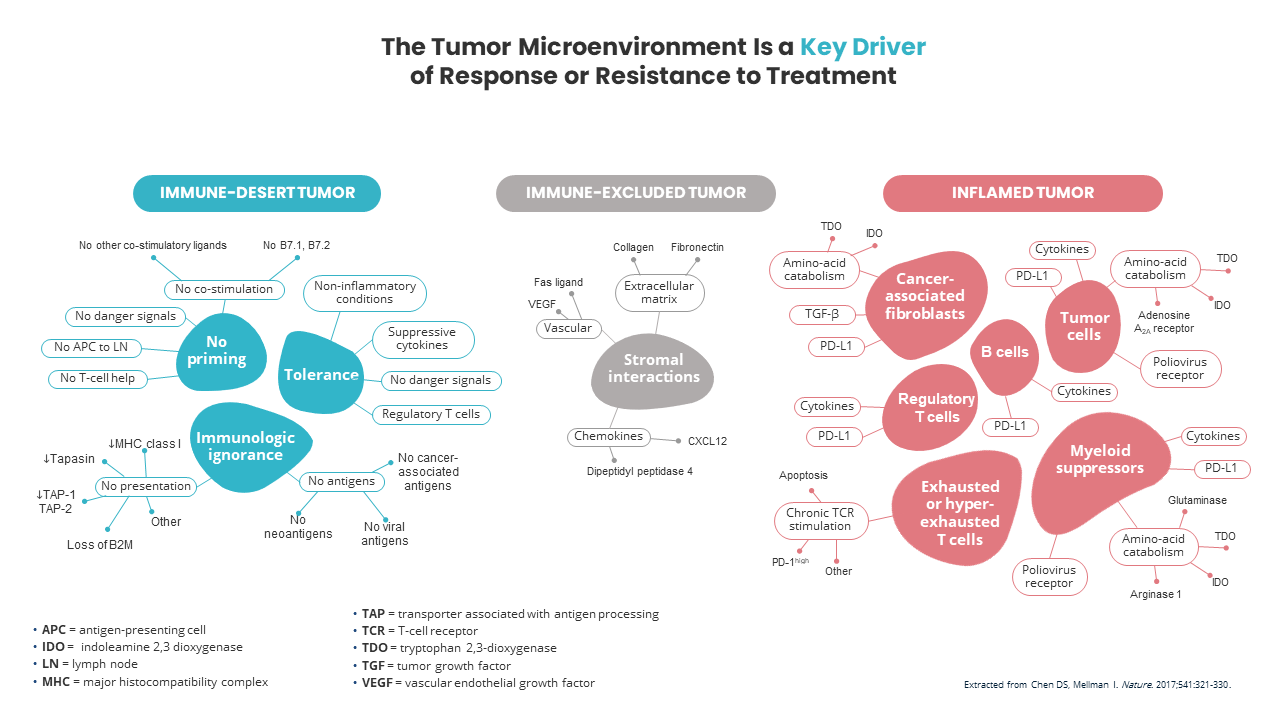
Immuno-oncology (IO) therapies have emerged as possible effective anti-cancer therapies for some patients. Immune checkpoint inhibitors have become a focus in IO research. They reduce T-cell inhibition, which allows the T-cells to target cancer cells. For patients that respond positively to this type of therapy, the benefits can last for years instead of just months with other types of therapies3. In addition, it is important to study cancer-immune phenotypes. Anticancer immunity in humans has been shown to separate into three main phenotypes: the immune-desert phenotype, the immune–excluded phenotype, and the inflamed phenotype4. Each of these cancer phenotypes has been associated with specific underlying biological mechanisms that could play a role in whether or not a person’s immune response can mount a response to cancer (Cancer-immune phenotypes [3: p. 324]) (Figure 1)4.
To minimize side effects and high treatment costs, choosing the best choice of immuno-therapy for an individual patient is critical. Understanding the immune TME can elucidate the biomarkers associated with response to immuno-oncology agents. Current candidates include PD-L1 expression, CD8+ tumor-infiltrating lymphocytes, tumor mutation load, neoantigen burden, and immune-related gene signatures5. Therefore, to investigate these multiple candidates simultaneously, multiplex immunohistochemistry (IHC) assays that examine the pharmacodynamic and spatial interactions of the TME are necessary5. In addition, validated and reproducible assays are needed. Furthermore, predictive tests to help oncologists identify the likelihood of response to monotherapy vs. combination therapy will have significant health and economic benefits5.
Discerning cell types using proteomics
Proteomics approaches have historically been used to distinguish between cell types. For example, IHC can be used to distinguish immune cell types and cancer subtypes such as HER2 expression in breast cancer2. In addition, the 2015 WHO classification of lung cancer introduced IHC in the classification schema. Thus, IHC is routinely used in clinical practice in diagnosing lung cancer and adenocarcinoma and squamous cell carcinomas can be distinguished with thyroid transcription factor 1 (TTF1) and p40 staining, respectively6.
New technologies in spatial biology
More recently, new technologies such as multiplexed immunofluorescence (mIF) are enabling the detection of molecular biomarkers and cancer subclones within their spatial context. These techniques, integrated with multi-omics data can generate a more profound understanding of inter- and intra-tumoral cell-to-cell variation and drive the next generation of research, diagnostic and therapeutic strategies in cancer research1.
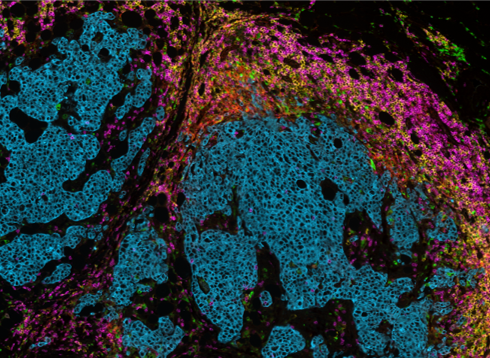
Lunaphore Technologies has developed an innovative, next-generation mIF technique using a microfluidic chip-based technology that standardizes and improves the traditional mIF processes (such as cyclic IF). Lunaphore has established a new approach, called sequential immunofluorescence (seqIF™) to perform automated, fast, and quantitative multiplex spatial proteomics without the pitfalls of cyclic IF (the need for directly-conjugated primary antibodies, complex upstream processing of antibodies, tissue-damaging signal removal steps and the inability to use standard, non-conjugated antibodies) (Figure 2). Multiplex staining with seqIF™ is an excellent method to visualize 40 antigens to quantitively detect spatial relationships that are functionally significant.
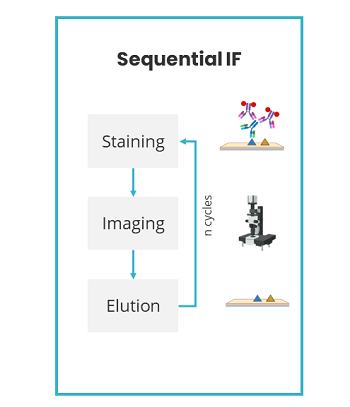
Spatial biology and mIF as the best method to assess response to treatment
Multiplex IF-based technologies provide many advantages in IO research including the detection of cell-to-cell spatial interactions that correlate with clinical outcomes and generate reliable and reproducible results in a high-throughput workflow7. A recent meta-analysis was performed by Johns Hopkins University on data pooled from more than 50 studies, on more than 10 different solid tumor types and over 8,000 patients8. Each study assessed the predictive value of one or more biomarker assays intended to determine the likelihood of response to anti–PD-1/PD-L1 therapy, the leading class of immunotherapy. The meta-analysis showed that the spatial biology revealed by mIHC or mIF, which included cellular protein co-expression, localization, and arrangement, correlated better with patient response than with the other approaches8.
Choosing the right technology for your spatial biology research
While mIF enables the study of multiple biomarkers and provides critical information on cell-cell interaction in the TME, these assays require robust validation and flexibility to be used across multiple tissue types. Lunaphore’s COMET™ instrument enables researchers to quickly develop and optimize multiplex panels by allowing them to use any commercially available antibodies while providing rapid, automated, high-throughput workflows.
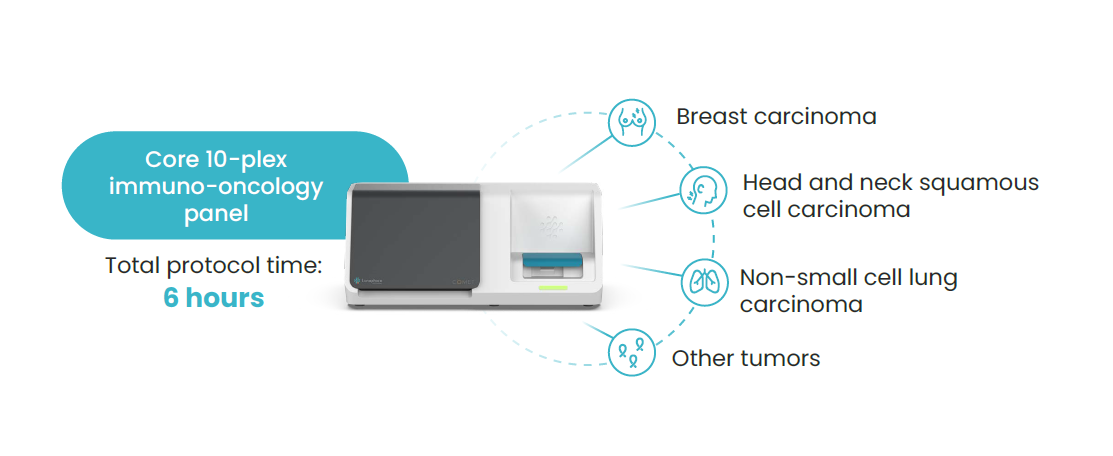
Lunaphore has demonstrated that an IO panel developed on COMET™ can be easily transferred from one tissue type to another, enabling researchers to identify core biomarkers across different tumors for the development of novel targeted immunotherapies (Figure 3).
References:
- Lewis SM, et al. Spatial omics and multiplexed imaging to explore cancer biology. Nat Methods. 2021 Sep;18(9):997-1012. doi: 10.1038/s41592-021-01203-6.
- Pan D, Jia D. Application of Single-Cell Multi-Omics in Dissecting Cancer Cell Plasticity and Tumor Heterogeneity. Front Mol Biosci. 2021 Oct 15;8:757024. doi: 10.3389/fmolb.2021.757024.
- Wilky BA. Immune checkpoint inhibitors: The linchpins of modern immunotherapy. Immunol Rev. 2019 Jul;290(1):6-23. doi: 10.1111/imr.12766.
- Chen DS, Mellman I. Elements of cancer immunity and the cancer-immune set point. Nature. 2017 Jan 18;541(7637):321-330. doi: 10.1038/nature21349.
- Mehnert JM, et al. The Challenge for Development of Valuable Immuno-oncology Biomarkers. Clin Cancer Res. 2017 Sep 1;23(17):4970-4979. doi: 10.1158/1078-0432.CCR-16-3063. PMID: 28864725; PMCID: PMC5657536.
- Yatabe Y, et al. Best Practices Recommendations for Diagnostic Immunohistochemistry in Lung Cancer. J Thorac Oncol. 2019 Mar;14(3):377-407. doi: 10.1016/j.jtho.2018.12.005.
- Hoyt CC. Multiplex Immunofluorescence and Multispectral Imaging: Forming the Basis of a Clinical Test Platform for Immuno-Oncology. Front Mol Biosci. 2021 Jun 2;8:674747. doi: 10.3389/fmolb.2021.674747.
- Lu S, et al. Comparison of Biomarker Modalities for Predicting Response to PD-1/PD-L1 Checkpoint Blockade: A Systematic Review and Meta-analysis. JAMA Oncol. 2019 Aug 1;5(8):1195-1204. doi: 10.1001/jamaoncol.2019.
Related Articles
Reproducibility and tissue integrity: spatial biology without compromises
Posted on 08 Apr 2022
Read PostAssay development: how to overcome the challenges of TSA-based mIHC assays
Posted on 11 Oct 2022
Read Post