Technical Note
Reproducibility and tissue integrity: spatial biology without compromises
Posted on:
The rise of multiplex spatial biology
Immunohistochemistry (IHC) is well-established in spatial biology research and diagnostics practices as a tool to assess tissue pathogenicity and potential therapies. One of its most popular techniques, immunofluorescence (IF), often used in tissue-based research, was once limited to labelling only one marker per tissue section1. However, since then, the need to further explore the tissue microenvironment in a multidimensional and interconnected manner has fueled multiple advances in IF applications. Presently, detection of two or more antigens on the same tissue is common and can be performed sequentially or simultaneously2. This is referred to as multiple labeling or multiplex IF (mIF)3. The use of quantitative multiplex analysis has spread through numerous research applications ranging from oncology, immunology, immunotherapy development, infectious diseases, neuroscience, and other pathological disease states4.
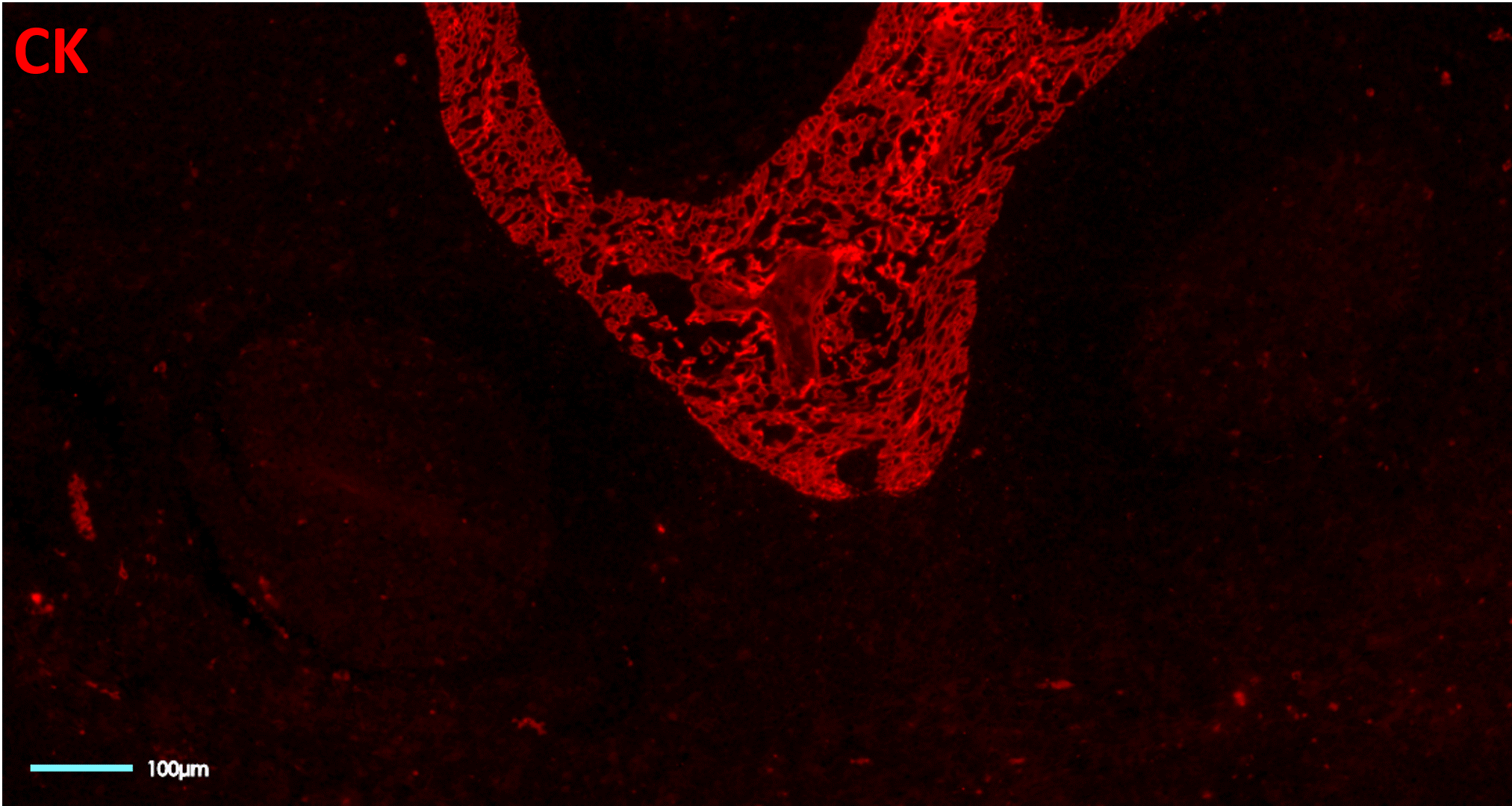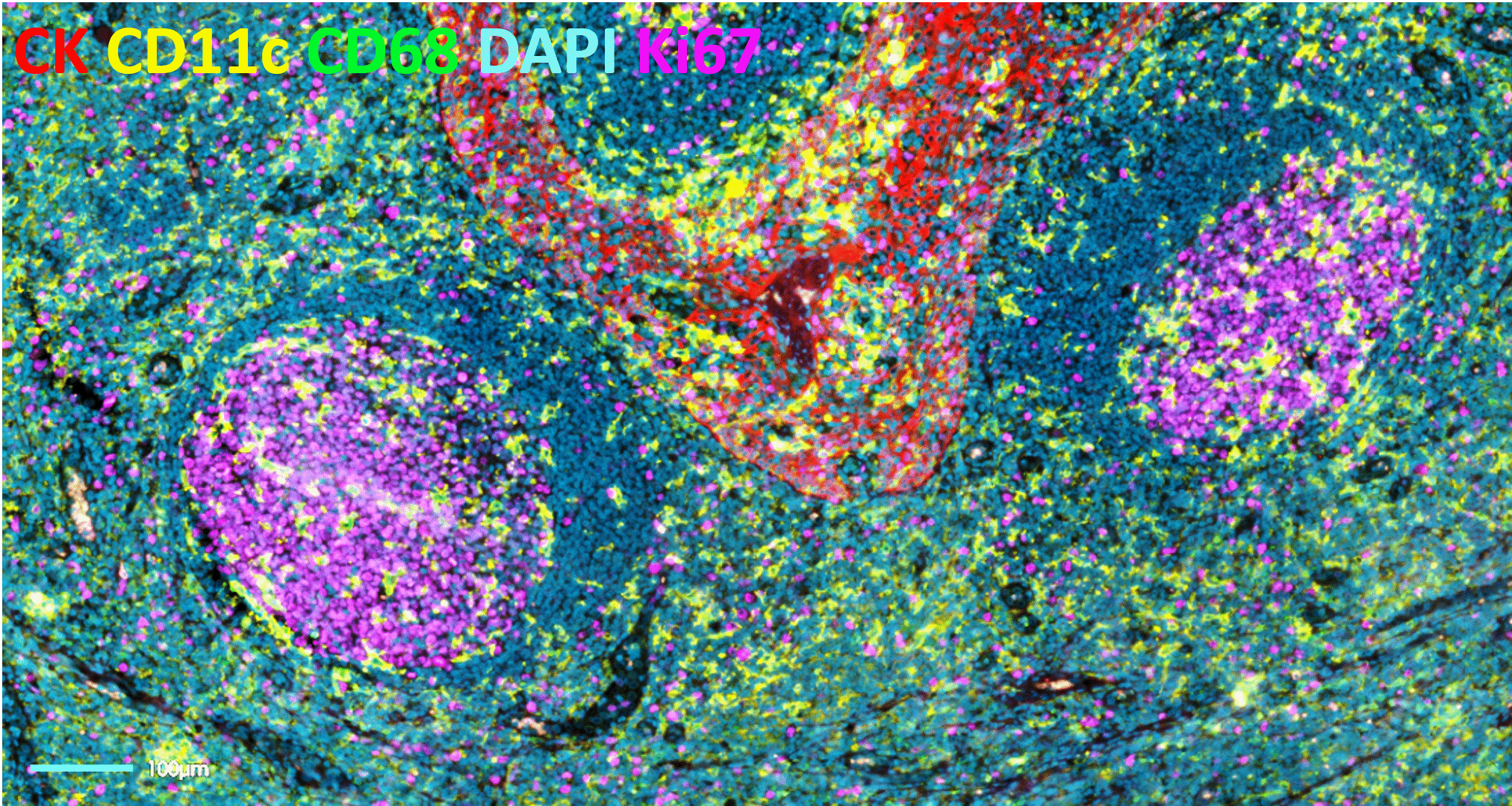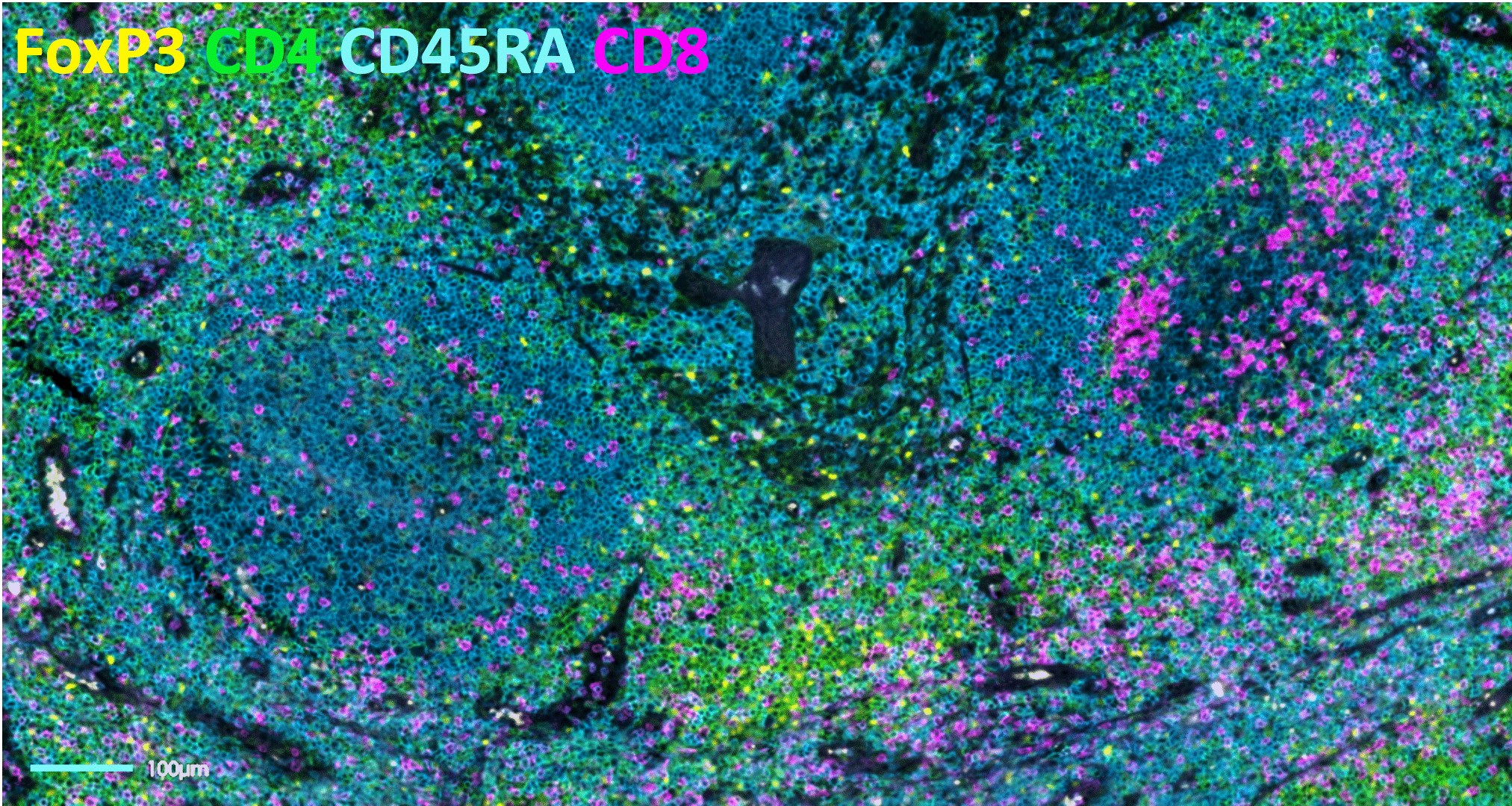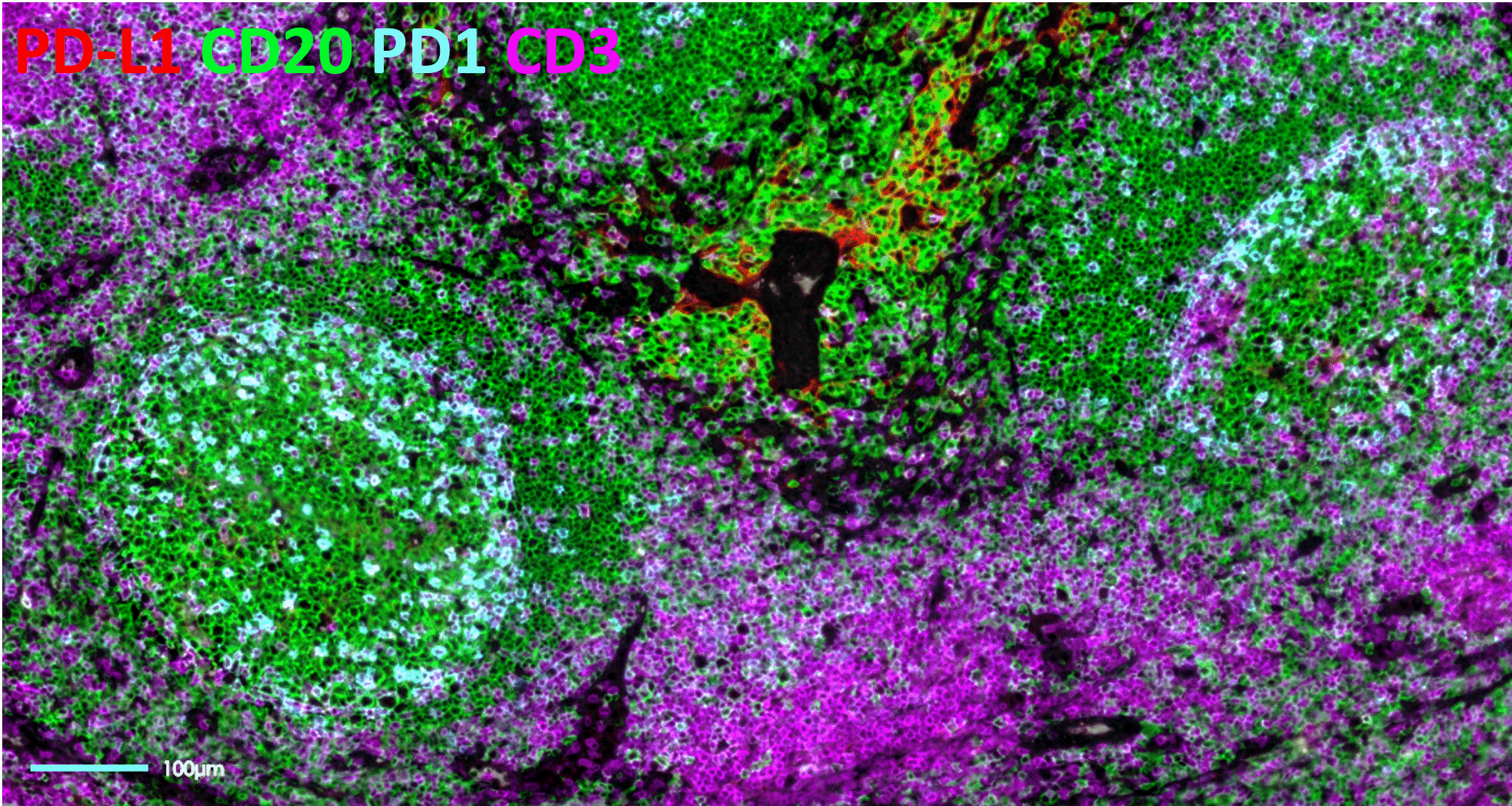
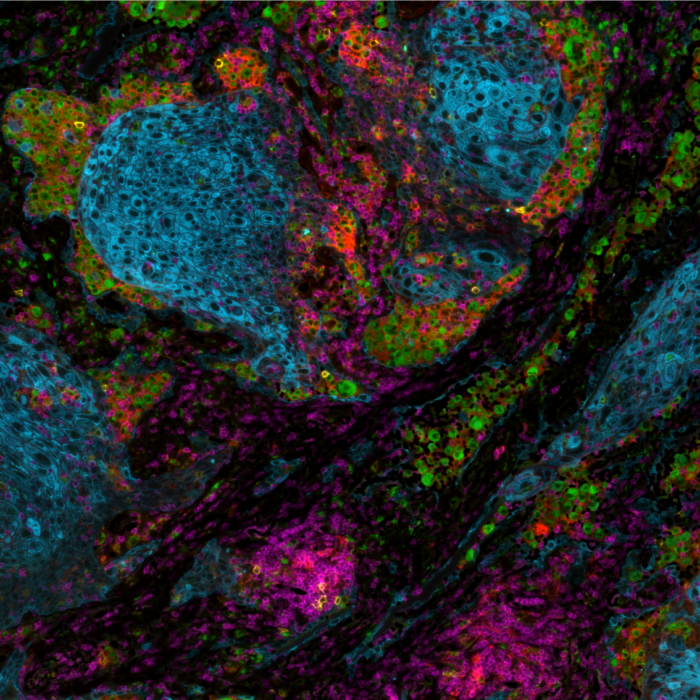
The bumpy road from multiplex to hyperplex spatial biology
Adoption of spatial biology applications is rapidly increasing, both in number and content depth. Fundamental discovery, as well as translational research laboratories, require the identification of an increasingly large number of markers on a single slide to obtain high-plex data, also known as hyperplex analysis. However, exsting and new technical approaches are required to overcome numerous technical challenges:
Tyramide Signal Amplification (TSA)-based staining is one of the most classic approaches to mIF. It involves the detection of all markers on a tissue sample simultaneously. It is, therefore, limited by the number of microscope channels or, in some cases, the spectral unmixing capabilities available5. For this reason, TSA-based mIF is rarely used above 10-plex analysis.
Cyclic IF is another well-known multiplexing method that aims at breaking through this barrier: it consists of repeated cycles of single-plex or low-plex staining, using antibodies that are directly conjugated to a fluorophore, followed by imaging and signal removal steps on the same tissue slide, generating individual images that can later be stacked together forming a hyperplex image6. Nevertheless, cyclic IF does come with significant challenges:
- Issues with tissue preservation: cyclic IF methods that require multiple rounds of signal removal by deactivation or strip-off of fluorescently-labeled antibodies, can use acidic buffers, strong reducing agents, proteases, heating steps or antibody deactivation through chemical processes or photobleaching (Figure 2). These methods raise concerns around tissue preservation, reproducibility, and quality of results.

- Technical limitations in plex level: repeated harsh cycles of signal removal present a technical barrier in the number of cycles that can be performed and, therefore, in the maximum number of markers that can be simultaneously analyzed without significant damage to the tissue integrity.
- Limited to use with primary-conjugated antibodies: laboratories using cyclic IF typically need to develop new assay protocols from scratch. Not being able to leverage existing validated libraries of non-conjugated antibodies, can be very time-consuming, taking up weeks, or, most often, months. Moreover, the absence of secondary antibodies in this approach prevents the use of any amplification, and detection of low expressing markers can be challenging.
Conjugation-based approaches require highly manual, complex, and time-consuming upstream conjugations or barcoding of primary antibodies (Figure 3). These steps often come with high unwanted variability, dramatically affecting the assay reproducibility, and conjugations themselves have a potential failure rate of up to 75%.
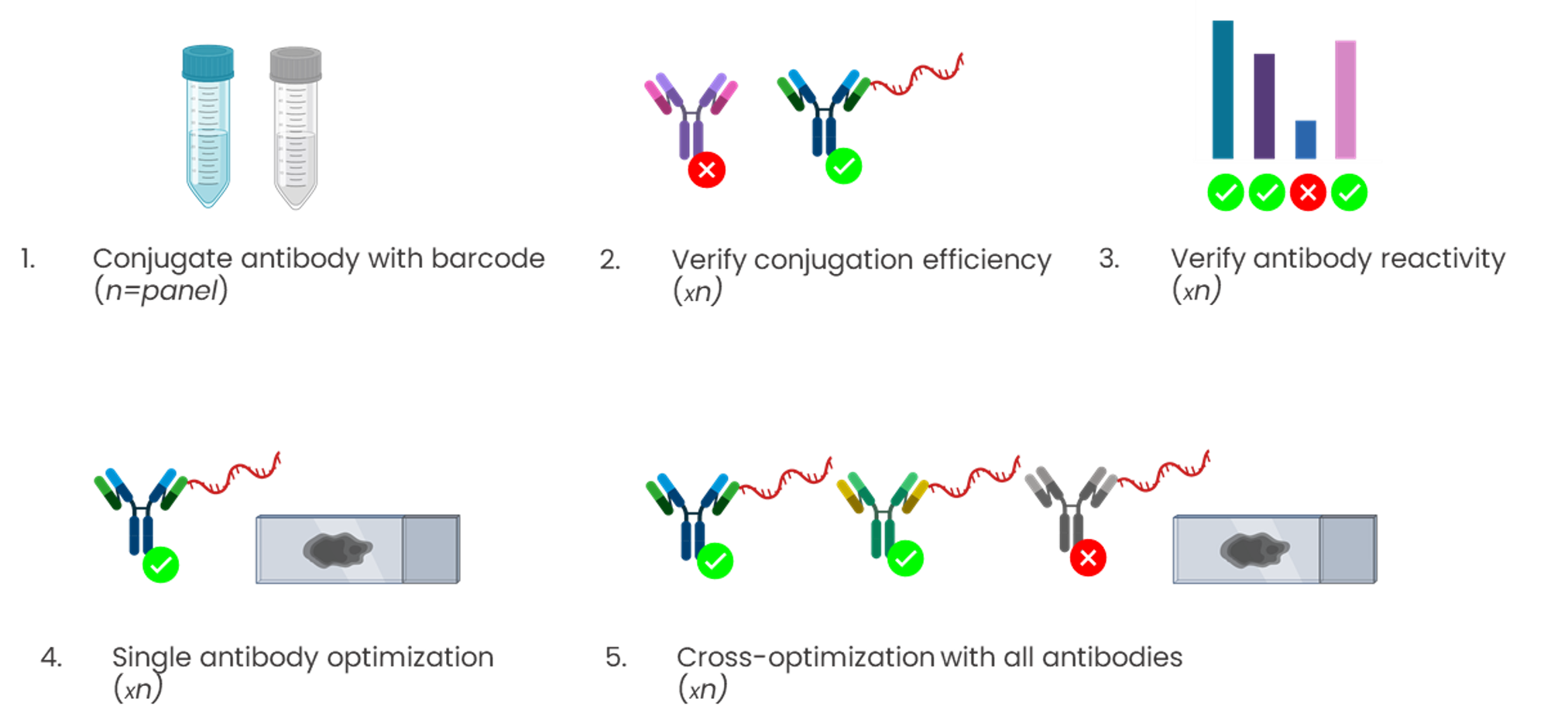
The elimination of hyperplexing barriers with sequential immunofluorescence (seqIF™)
A next-generation mIF technique is a microfluidic chip-based technology that standardizes and improves the cyclic IF process, performing automated, fast, quantitative multiplex spatial proteomics without the pitfalls of cyclic IF (the need for directly-conjugated primary antibodies, complex upstream processing of antibodies, tissue-damaging signal removal steps and the inability to use non-conjugated antibodies). This efficient method developed by Lunaphore is called sequential immunofluorescence (seqIF™). Multiplex staining with seqIF™ is an excellent method to visualize up to 40 antigens to explore spatial relationships that are functionally significant3,8.
This method relies on three key variations from cyclic IF:
- Outstanding tissue preservation and reproducibility with FFeX technology: the seqIF™ method relies on a novel precision-fluidics Imaging Chip that leverages the properties of the Fast-Fluidic Exchange Technology (FFeX), an innovation which significantly reduces reaction times down to a few minutes (Figure 4). The system performs repeated cycles of staining, imaging, and elution (an antibody removal step). Following the staining step, images of the stained area are acquired. The short exposure time of tissues to harsh reagents preserves the integrity of the tissue throughout numerous cycles. Lunaphore’s FFeX technology enables fine control of all conditions ensuring robust assays and highly reproducible results.

- Use of standard, non-conjugated antibodies: seqIF™ uses established species-specific secondary antibodies conjugated with a fluorescent tag, which enables secondary-antibody linear amplification. More importantly, it allows researchers to leverage existing libraries of validated primary antibodies without the need for any complex upstream processing.
- A gentle, elution-based antibody removal: following staining and imaging, a gentle antibody stripping step is performed, avoiding damage to the tissue. This stripping is called elution and it benefits from innovations in both its chemistry, with the use of a dedicated elution buffer; and the reaction’s thermodynamic profile, leveraging a patented microfluidic incubation inside the Imaging Chip with no-static wash, a mild temperature step, and a very short duration, around 2 minutes instead of several hours (Figure 5).

seqIF™ on COMET™, the only high-throughput, hyperplex solution using non-conjugated antibodies
The seqIF™ method runs on Lunaphore systems. The COMET™ platform, a powerful discovery and translational research tool, can be used to easily assess 40 markers on the same tissue (Figure 6). With a throughput of 20 slides of 20-plex per week, it enables scientists to substantially scale up their research capabilities.
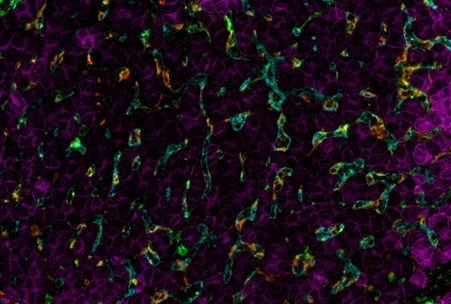
Assessing the best staining conditions and using a gentle but efficient antibody removal process is necessary after each staining round. In the COMET™ platform, built-in characterization protocol templates guide the user step-by-step in the assessment of the staining quality for a single marker, or a mix of two markers, simultaneously, for elution efficiency and for epitope stability.
Precision-fluidics-based seqIF™ on COMET™ clearly demonstrates high epitope stability and elution efficiency over multiple cycles of antibody staining and stripping, in both Formalin-Fixed, Paraffin-Embedded samples (FFPE) and frozen tissue. The robustness of the staining quality provided by COMET™ was proven across 15 different tissue types for more than 50 markers up to 20 cycles.
Test the tissue integrity of your samples with seqIF™ on COMET™
If you are interested in assessing the seqIF™ impact on the tissue integrity of your samples, or other questions related to COMET™ performance in your laboratory, check out the Access Lab to determine the best fit for your experimental needs.
References:
- Tan WCC, et al. Overview of multiplex immunohistochemistry/immunofluorescence techniques in the era of cancer immunotherapy. Cancer Commun (Lond). 2020;40(4):135-153. doi:10.1002/cac2.12023
- Pirici D, et al. Antibody elution method for multiple immunohistochemistry on primary antibodies raised in the same species and of the same subtype. J Histochem Cytochem. 2009 Jun;57(6):567-75. doi: 10.1369/jhc.2009.953240.
- Bolognesi MM, et al. Multiplex Staining by Sequential Immunostaining and Antibody Removal on Routine Tissue Sections. J Histochem Cytochem. 2017 Aug;65(8):431-444. doi: 10.1369/0022155417719419.
- Migliozzi D, et al. Microfluidics-assisted multiplexed biomarker detection for in situ mapping of immune cells in tumor sections. Microsyst Nanoeng. 2019 Nov 6;5:59. doi: 10.1038/s41378-019-0104-z.
- Pivetta E, et al. Multiplex staining depicts the immune infiltrate in colitis-induced colon cancer model. Sci Rep. 2019 Sep 2;9(1):12645. doi: 10.1038/s41598-019-49164-3.
- Lin JR, et al. Highly multiplexed imaging of single cells using a high-throughput cyclic immunofluorescence method. Nat Commun. 2015 Sep 24;6:8390. doi: 10.1038/ncomms9390.
- Wählby C, et al. Sequential immunofluorescence staining and image analysis for detection of large numbers of antigens in individual cell nuclei. Cytometry. 2002 Jan 1;47(1):32-41.
Related Articles
Assay development: how to overcome the challenges of TSA-based mIHC assays
Posted on 11 Oct 2022
Read Post