Poster
Putting the pieces together: the spatial evolution of cancer research
Posted on:
Acknowledging and integrating the inherent heterogeneity of cancers is essential for the advancement of cancer research and the personalization of treatment modalities. The diverse functional and structural characteristics of cancers, which vary significantly across patients, present a formidable challenge in oncology, impacting treatment efficacy and prognosis. Cancer research has journeyed through an extensive and fascinating history, evolving from the high-level understanding of tumors to the sophisticated, detailed analyses we conduct today.
Evolving perspectives in cancer research
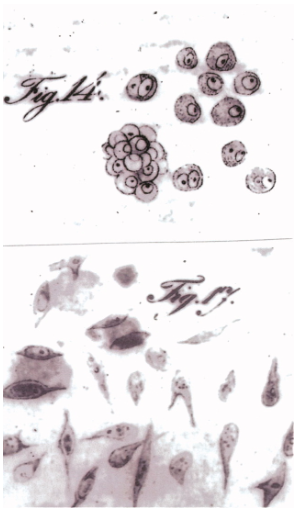
In the early 19th century, Johannes Peter Müller, a pioneering German scientist, laid the groundwork for cancer research through his initial descriptions of cancer cells1 (Figure 1). Müller’s seminal contributions marked the beginning of an era of significant breakthroughs, progressively demystifying the complexities of cancer.
In the 20th century, a significant development in cancer research was the introduction of immunohistochemistry (IHC). This technique helped identify particular tissue features and protein targets within tumor cells, transforming the way cancer is studied. IHC paved the way for more advanced characterizations of cancer by providing crucial insights into the cellular changes that cause cancer development and progression. The introduction of multiplex immunofluorescence (mIF) met the demand for advanced analytical methods by enabling the simultaneous detection of multiple markers on a single tissue section. This advancement significantly enhanced the ability to analyze the molecular complexities of the tumor microenvironment (TME). The revolution in single-cell technologies represents the latest frontier in cancer research, offering even more granular insights into cancer heterogeneity2. By enabling precise analysis at the individual cell level, these technologies are refining our understanding of the TME’s dynamics, further empowering researchers to identify novel avenues for intervention.
Grasping the spatial dynamics of the TME with integrated RNA and protein profiling
Today, spatial multiomics, which involves the simultaneous analysis of different types of molecules within their native spatial context, is revolutionizing cancer research. This technique enables the exploration of tumor heterogeneity at a molecular level, all while preserving the spatial context crucial for a comprehensive understanding of tumor biology. By delineating the molecular profiles of both cancerous and non-cancerous cells within the TME and tracking how these profiles vary across different spatial regions of the tumor, researchers can unearth the underlying mechanisms that drive tumor growth, metastasis, and resistance to treatments.
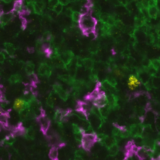
A recent study presented at the Advances in Genome Biology and Technology (AGBT) Society General Meeting 2024 highlighted a novel same-section, fully automated spatial multiomics approach on the COMET™ platform for comprehensive TME characterization. The workflow combines the RNAscope™ HiPlex Pro Assay3 with sequential immunofluorescence (seqIF™)4. Utilizing an optimized target retrieval protocol and protease-free tissue pre-treatment, this fully automated multiomics workflow supports the concurrent detection of desired RNA and protein targets (Figure 2).
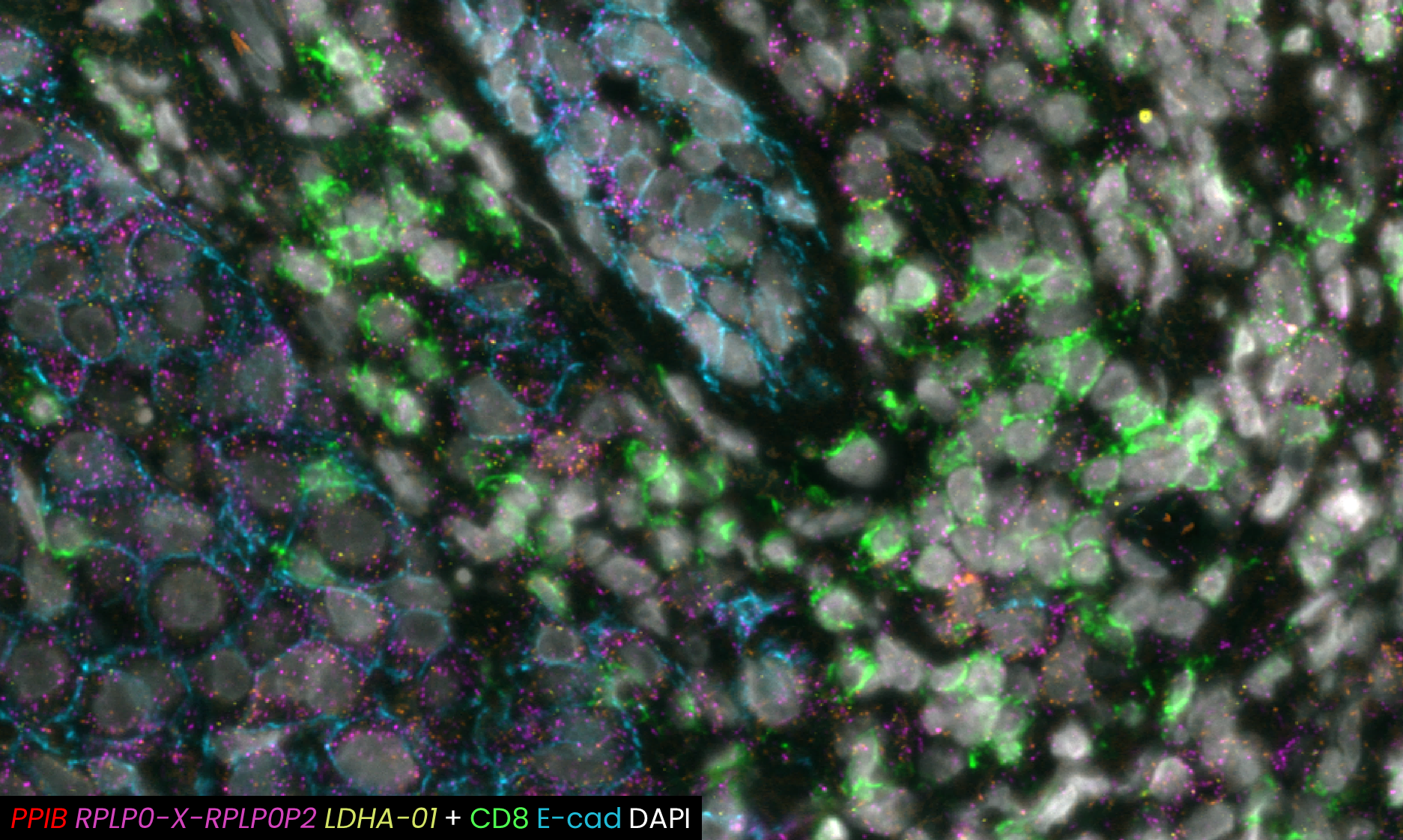
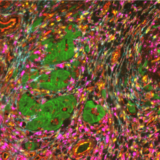
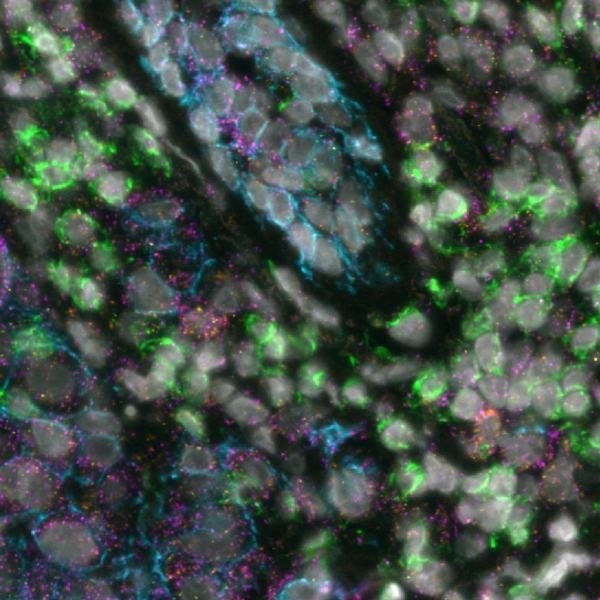
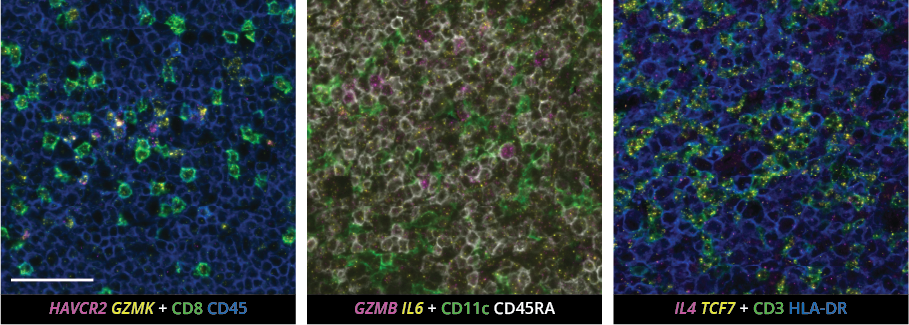
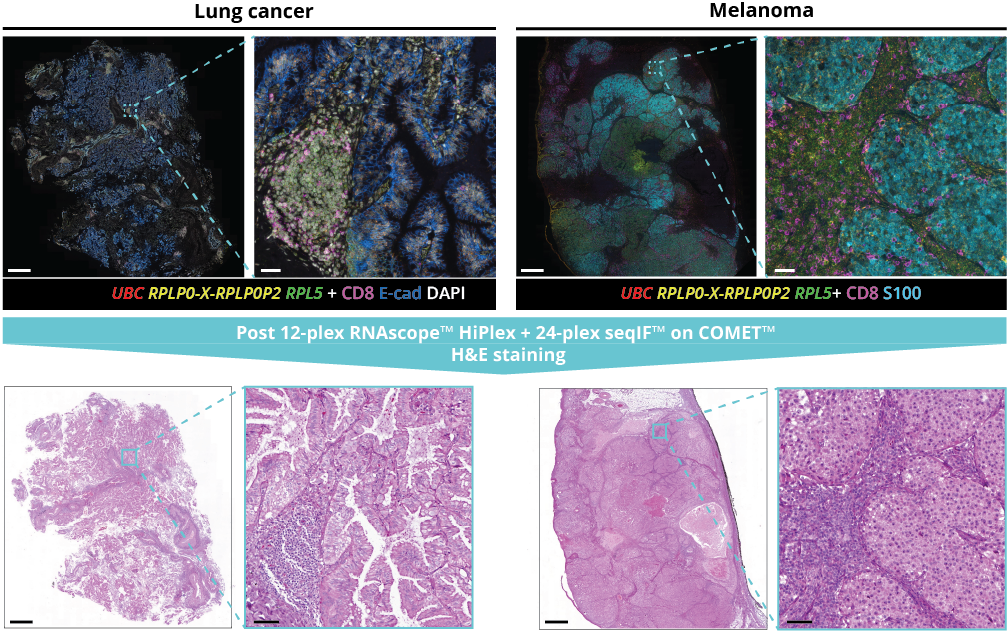
What makes the COMET™ platform particularly remarkable is its full automation capability, which significantly streamlines the research process, reducing the potential for human error, thereby increasing assay throughput and reproducibility. The seamless RNA and protein targets profiling enables enhanced comprehension of immune cell spatial organization and facilitates the characterization of their activation states (Figure 3). Furthermore, following analysis on the COMET™ platform, tissues are fully preserved. The same slide can be used to perform any additional downstream modalities, such as H&E (Figure 4). The COMET™ platform exemplifies the desirable features propelling spatial multiomics technology forward, offering researchers unparalleled opportunities to explore the complexities of the tumor microenvironment and beyond with exceptional detail and accuracy.
As we continue to harness the power of spatial multiomics, we stand on the brink of a new era in cancer research—one where our growing comprehension of the tumor microenvironment leads to more effective, targeted treatments and ultimately, better outcomes for patients. The journey from the foundational work of pioneers like Johannes Müller to today’s sophisticated spatial analyses highlights not only how far we’ve come but also the promising direction in which we’re headed. With these advancements, the future of cancer research and treatment looks brighter than ever, promising a deeper understanding and more effective combat against this complex disease.
- discover the same-section, fully automated workflow on COMET™ combining RNAscope™ and seqIF™;
- explore the TME molecular landscape with 12-plex RNA plus 24-plex protein multiomics panel;
- uncover advanced imaging capabilities on COMET™ including the acquisition of z-stacks, followed by automated maximum intensity projection (MIP);
- assess tissue preservation across multiple tissue types by hematoxylin and eosin (H&E) staining after same-section multiomics protocol.
References:
- Hajdu S. A note from history: Landmarks in history of cancer, part 3. Cancer. 2012. 118(4):1155-68. doi: 10.1002/cncr.26320.
- Lewis S. et al. Spatial omics and multiplexed imaging to explore cancer biology. Nat Methods. 2021. 18(9):997-1012. doi: 10.1038/s41592-021-01203-6.
- Wang F. et al. RNAscope: A Novel In situ RNA Analysis Platform for Formalin-Fixed Paraffin-Embedded Tissues. J Mol Diagn. 2012. 14(1):22-9. doi: 10.1016/j.jmoldx.2011.08.002.
- Rivest F. et al. Fully automated sequential immunofluorescence (seqIF) for hyperplex spatial proteomics. Sci Rep. 2023. 13(1):16994. doi: 10.1038/s41598-023-43435-w.
Related Articles
Cytokines, spatial activation and tumor infiltration: the future of immunotherapy
Posted on 13 Dec 2023
Read Post