Spatial Biology Education
Spatial biology meets Candida albicans: deciphering infections, immune interactions, and therapeutic insights
Posted on:
Candida albicans is a common fungal pathogen that can lead to various infections, ranging from mucosal infections to life-threatening sepsis1. Despite advances in modern medicine, the mortality rate associated with Candida albicans sepsis remains high1. This has spurred a great deal of research focusing on understanding disease mechanisms and developing novel therapies. Spatial biology research is revolutionizing how we perceive and analyze complex biological systems. Unlike conventional methods that might overlook single-cell approaches, spatial biology focuses on preserving the structural context of cells and tissues. This approach allows researchers to investigate the distribution pattern of molecules, cellular interactions, and microenvironmental influences. In the context of Candida albicans, spatial biology enables researchers to analyze the interplay between this fungus and its surroundings closely.
Pioneering progress in Candida albicans sepsis research
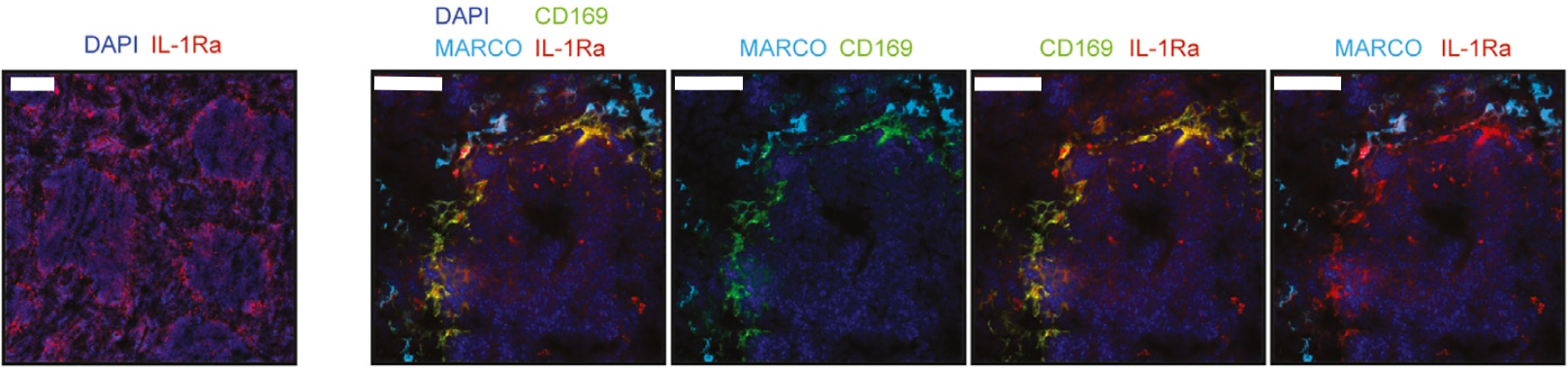
Recently, a research paper published by Gander-Bui et al. facilitated by the LabSat® platform has expanded our understanding of Candida albicans‘ infectious cycle, offering new therapeutic routes to limit its burden2. Authors demonstrated that the interleukin-1 receptor antagonist (IL-1Ra) secreted by macrophages served as a natural immune checkpoint, hindering the effective clearance of pathogens. The IL-1Ra protein is secreted into the bloodstream by CD169+ macrophages in the splenic marginal zone. Conversely, MARCO+ macrophages were observed to lack IL-1Ra expression (Figure 1). Notably, the focused elimination of this molecule resulted in enhanced protection against fatal Candida albicans sepsis.
Introducing sequential immunofluorescence (seqIF™): a breakthrough in multiplex protein detection
The study by Gander-Bui et al. used sequential immunofluorescence (seqIF™) on LabSat® to identify CD169+ macrophages of the splenic marginal zone as main producers of IL-1Ra. SeqIF™ presents a novel and straightforward method to detect numerous proteins in a single tissue section with unparalleled efficiency.