Poster
Beyond the limits of current checkpoint blockade approaches: how spatial biology tools can help unlock cancer therapeutics
Posted on:
The interplay among tumor cells, the surrounding tissue, and infiltrating innate and adaptive immune cells create a distinct environment that varies depending on the tumor type, stage, host, and treatment history1. This complex ecosystem is called the tumor microenvironment (TME) and refers to the surrounding cellular and non-cellular components within a tumor core and its periphery. It includes various tissue cell types, an extracellular matrix, blood vessels, immune cells, fibroblasts, and signaling molecules that enable communication within the TME and between the TME and the rest of the body.
The tumor-immune microenvironment presents a complex and challenging landscape for cancer therapeutics.
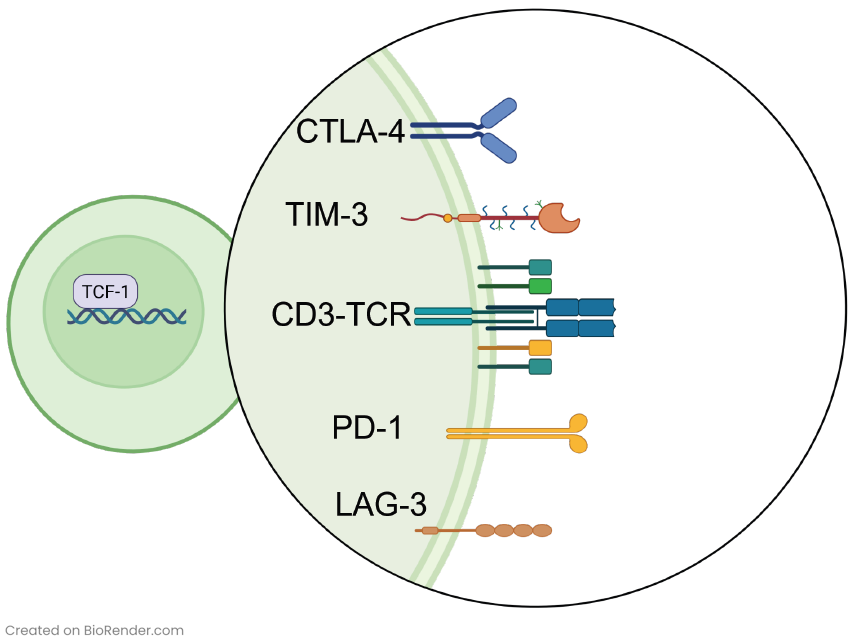
The identification and understanding of immune checkpoint proteins that regulate immune responses have revolutionized the field of cancer care. Immune checkpoints serve as “brakes” on the immune system, preventing overactivation and potential damage to healthy tissues. They help maintain self-tolerance and protect against autoimmunity. However, cancer cells hijack these safety mechanisms to bypass immune recognition. Well-known immune checkpoint proteins include Programmed Death-1 (PD-1) and Programmed Death Ligand-1 (PD-L1): PD-1 is expressed on the surface of activated T cells (Figure 1), while PD-L1 is often upregulated on tumor cells and immune cells within the TME. When PD-1 on T cells binds to PD-L1 on tumor cells, it inhibits T cell activity and prevents effective anti-tumor immune responses.
Immune checkpoint inhibitors have become a relevant tool in cancer immunotherapy. In 2011, the FDA approved ipilimumab, the first immune checkpoint targeting agent that specifically binds cytotoxic T-lymphocyte-associated protein 4 (CTLA-4). This pivotal development marked the emergence of immune checkpoint inhibitor (ICI) therapy. Since then, additional monoclonal antibodies (mAbs) blocking checkpoint proteins like PD-1 or PD-L1 have received FDA approvals2. While subsets of patients have exhibited remarkable and long-lasting clinical responses, it is important to acknowledge that most patients do not respond to ICI therapy today. Additionally, part of the patients that display a positive initial response to ICI therapy may develop resistance to treatment within a few months2. To effectively harness the immune system for inhibiting tumor progression, it is imperative to have a comprehensive understanding of the distribution, function, and interaction of the immune cells within the TME. This critical knowledge can help researchers and clinicians develop novel strategies to optimize immune checkpoint blockade efficiency and improve patient outcomes.
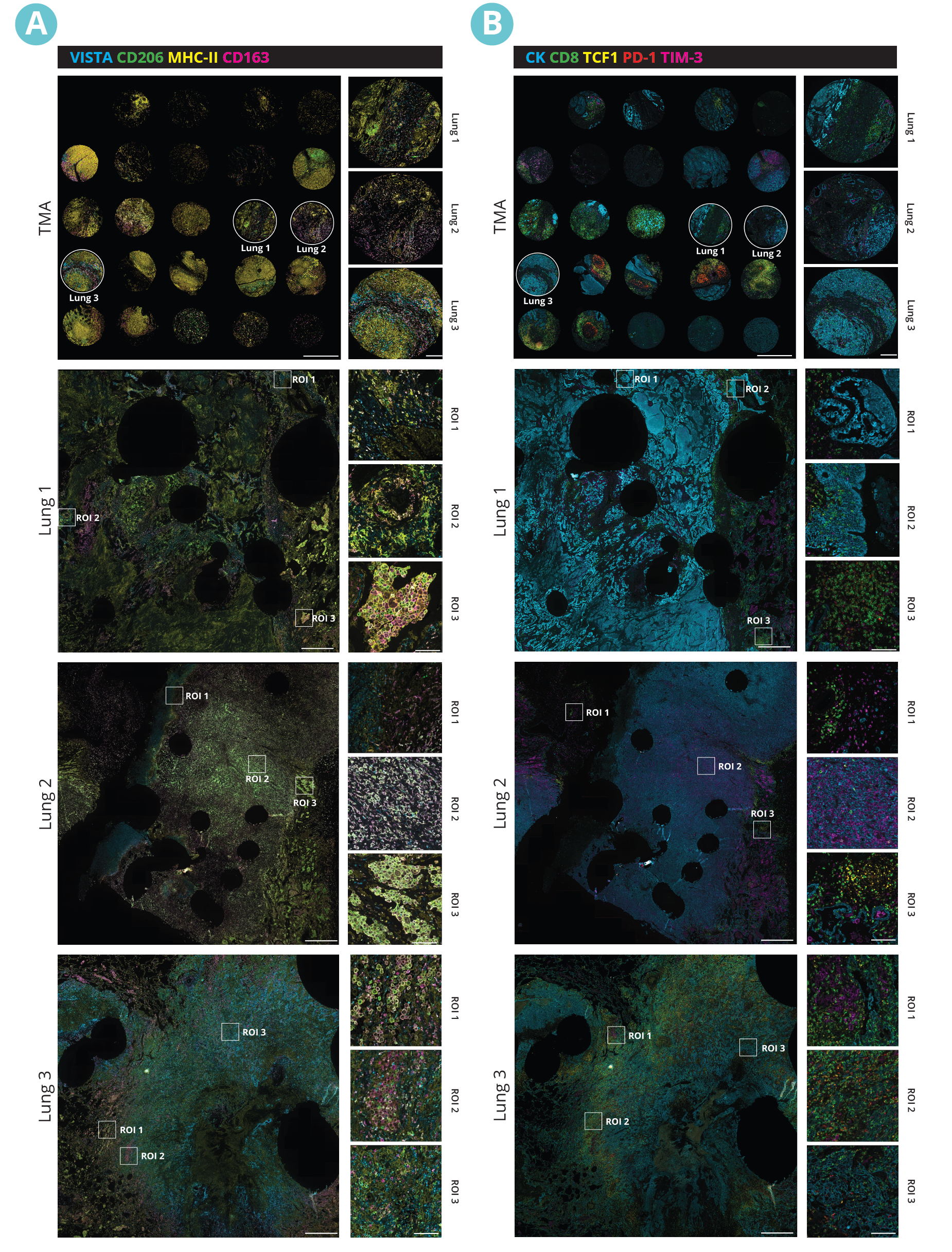
Multiplex immunofluorescence (mIF) reveals the heterogeneity of the TME
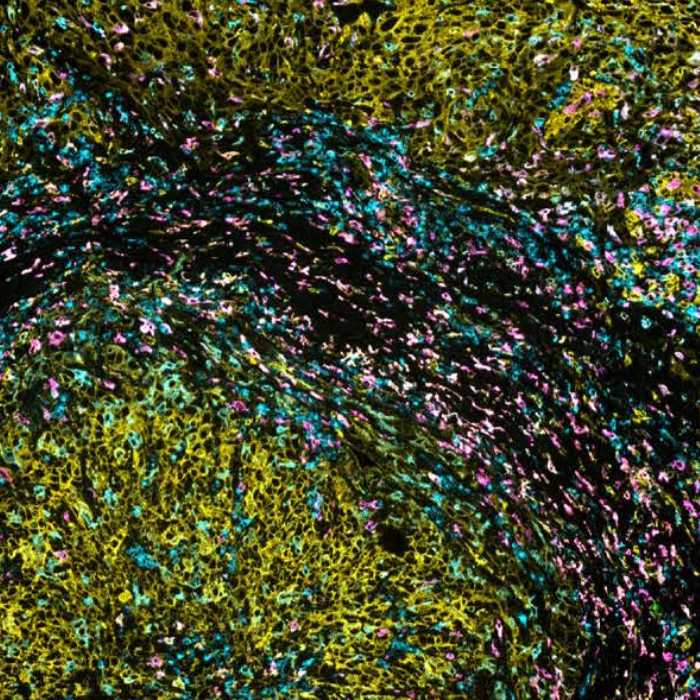
Spatial proteomics technologies have provided valuable insights into the heterogeneity of the tumor-immune microenvironment. By analyzing individual cells within the TME, mIF has revealed the presence of diverse immune cell populations, each exhibiting unique characteristics and functions3. In a recent study exhibited at the 2023 Annual Congress of the European Association for Cancer Research (EACR 2023), Lunaphore with Cell Signaling Technology (CST) presented a novel hyperplex panel allowing for in-depth characterization of the immune compartment in the context of the TME with a focus on immune checkpoint biomarkers. In this case study, slides with human lung tumor FFPE tissue slices were stained on COMET™. A 22-plex panel was developed and composed of a 13-plex SPYRE™ Immuno-Oncology Core Panel (CD3, CD4, CD8, CD11c, CD20, CD45, CD56, CD68, αSMA, FOXP3, PD-1, PD-L1, Ki-67), expanded with Cytokeratin as a carcinoma marker and eight additional antibodies from CST (CD163, CD206, CTLA-4, MHC-II, LAG-3, TCF1, TIM-3, and VISTA). This optimized panel can be used in automated workflow on COMET™ in various tissue types and allows for deep interrogation of the TME (Figure 2).
The sequential immunofluorescence (seqIF™) protocol offers a powerful solution for designing and executing off-the-shelf, same-species antibody panels without needing antibody labeling. The COMET™ platform can unlock spatial data, enabling in-depth sample analyses at the single-cell level.