Poster
Pioneering RNA and protein detection in the tumor microenvironment (TME): an automated same-slide multiomics
Posted on:
Spatial biology stands at the forefront of life sciences, offering a transformative approach to studying the intricate tumor microenvironment (TME). Unlike traditional methods, spatial biology reveals the localization and proximity of different cells to one another. This level of detail is crucial in understanding the dynamic interplay between tumor cells, immune cells, and the extracellular matrix, giving researchers insights into how these spatial interactions can influence tumor growth, metastasis, and response to therapies. Recent advancements in multimodal omics technologies have enabled scientists to define the TME at single-cell resolution, offering a unique chance to grasp the complexity of the TME. By mapping cells in their spatial context, this discipline reveals not only the individual characteristics of cells but also their relationships and interdependencies with neighboring cells and the surrounding extracellular matrix. The simultaneous observation of RNAs and proteins enhances the depth and breadth of molecular research and has profound implications for translational medicine and therapeutic innovations. The significance of examining the expression of numerous markers with spatial context has driven the creation of various innovative methods to enhance multiplexing potential. This blog explores the pioneering study presented at the Society for Immunotherapy of Cancer’s (SITC) 38th Annual Meeting, which combines the detection of RNA and protein on the same slide, all harmoniously orchestrated on the state-of-the-art COMET™ platform.
Bridging RNA and protein detection
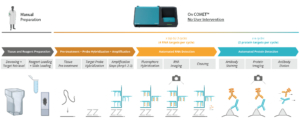
RNAscope™ is an in situ hybridization (ISH) assay for detecting RNA in various sample types1 and known for its high sensitivity and specificity. The RNAscope™ HiPlex allows for the simultaneous detection of up to 12 RNA targets in a single tissue section, preserving the spatial context. The sequential immunofluorescence (seqIF™) method detects protein biomarkers on virtually any tissue type. It consists of repeated staining cycles, imaging, and elution steps of antibody complexes while maintaining tissue antigenicity and morphology2. The study’s primary objective was to integrate the RNAscope HiPlex assay with the seqIF™ protocol, facilitating the concurrent detection of RNA and protein biomarkers in the same tissue section in one fully automated workflow (Figure 1).
A symphony of multiomics analysis
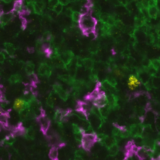
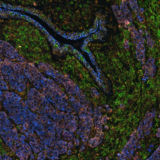
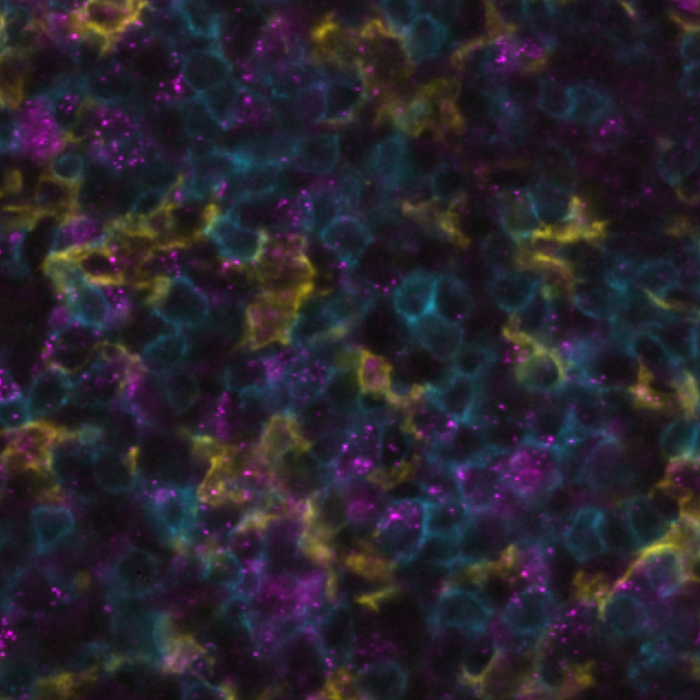
This integration allowed the development of a protocol enabling three cycles of RNA detection followed by consecutive cycles of seqIF™. The fully automated process on the COMET™ platform seamlessly performs all protocol steps, including tissue pre-treatment and probe hybridization, without any user intervention. By combining RNA and protein co-detection, researchers were able to look into intricate co-expression patterns and reveal a molecular map of the TME (Figure 2). In detail, the research showed an in-depth analysis of subclasses of cytokine-secreting CD3+ T-cell lymphocytes and CD56+ Natural Killer cells expressing GZMB, together with a characterization of tumor cells and their secretome (e.g., IL6+ cancer cells in lung cancer).
Related Articles
Cytokines, spatial activation and tumor infiltration: the future of immunotherapy
Posted on 13 Dec 2023
Read Post